George E. Jaskiw1,2, Mark E. Obrenovich3,4,5, Sirisha Kundrapu2, Curtis J. Donskey2,6
1Psychiatry Service, Veterans Affairs Northeast Ohio Healthcare System (VANEOHS), Cleveland, Ohio
2School of Medicine, Case Western Reserve University, Cleveland, Ohio
3Pathology and Laboratory Medicine Service, VANEOHS, Cleveland, Ohio
4Research Service, VANEOHS, Cleveland, Ohio
5Department of Chemistry, Case Western Reserve University, Cleveland, Ohio
6Geriatric Research, Education and Clinical Center, VANEOHS, Cleveland, Ohio
George E. Jaskiw
Psychiatry Service
116(A), VA Northeast Ohio Healthcare System
10701 East Blvd., Cleveland, OH 44106
Phone: 216-791-3800
Fax: 216 229-2396
gxj5@case.edu
10.20411/pai.v5i1.394
Jaskiw GE, Obrenovich ME, Kundrapu S, Donskey CJ. Changes in the Serum Metabolome of Patients Treated With Broad-Spectrum Antibiotics. Pathogens and Immunity. 2021;6(1):382-418. PubMed PMID. doi: 10.20411/pai.v5i1.394
Background: The gut microbiome (GMB) generates numerous small chemicals that can be absorbed by the host and variously biotransformed, incorporated, or excreted. The resulting metabolome can provide information about the state of the GMB, of the host, and of their relationship. Exploiting this information in the service of biomarker development is contingent on knowing the GMB-sensitivity of the individual chemicals comprising the metabolome. In this regard, human studies have lagged far behind animal studies. Accordingly, we tested the hypothesis that serum levels of chemicals unequivocally demonstrated to be GMB-sensitive in rodent models would also be affected in a clinical patient sample treated with broad spectrum antibiotics.
Methods: We collected serum samples from 20 hospitalized patients before, during, and after treatment with broad-spectrum antibiotics. We also collected samples from 5 control patients admitted to the hospital but not prescribed antibiotics. We submitted the samples for a non-targeted metabolomic analysis and then focused on chemicals known to be affected both by germ-free status and by antibiotic treatment in the mouse and/or rat.
Results: Putative identification was obtained for 499 chemicals in human serum. An aggregate analysis did not show any time x treatment interactions. However, our literature search identified 10 serum chemicals affected both by germ-free status and antibiotic treatment in the mouse or rat. Six of those chemicals were measured in our patient samples and additionally met criteria for inclusion in a focused analysis. Serum levels of 5 chemicals (p-cresol sulfate, phenol sulfate, hippurate, indole propionate, and indoxyl sulfate) declined significantly in our group of antibiotic-treated patients but did not change in our patient control group.
Conclusions: Broad-spectrum antibiotic treatment in patients lowered serum levels of selected chemicals previously demonstrated to be GMB-sensitive in rodent models. Interestingly, all those chemicals are known to be uremic solutes that can be derived from aromatic amino acids (L-phenylalanine, L-tyrosine, or L-tryptophan) by anaerobic bacteria, particularly Clostridial species. We conclude that judiciously selected serum chemicals can reliably detect antibiotic-induced suppression of the GMB in man and thus facilitate further metabolome-based biomarker development.
Keywords: serum, metabolome, antibiotic, L-tyrosine, L-phenylalanine, L-tryptophan, Clostridia
The gut microbiome (GMB) generates numerous small (< 2,000 Da) chemicals that can be absorbed, biotransformed, and either incorporated or excreted by the host [1-3]. The levels of such chemicals, their relationships, and temporal patterns can offer information about the state of the GMB, the gut wall, the liver, or other participating systems [4, 5]. GMB-derived metabolites may themselves be bioactive [6]. Thus, the characterization of their chemical profiles may have a wide range of diagnostic, prognostic, or therapeutic applications [4, 5].
Development of such potential biomarkers requires identifying the GMB-sensitive constituents of the metabolome. While germ-free mice and rats (GF) have proved invaluable in this regard [7], they have no true human homologue. An alternative approach is to evaluate antibiotic regimens that in rodent models selectively suppress certain microbial taxa [8-11] or eliminate most of the GMB, thus generating a pseudo-GF status [12]. Although it has been more than 65 years since antibiotic treatment was first observed to lower excretion of volatile phenols in man [13], a current literature search identified only 5 studies of antibiotic effects on the human serum metabolome [14-18]. Given the high and rising global use of antibiotics [19], the paucity of metabolomic studies in antibiotic-exposed humans constitutes a striking knowledge gap that must be acknowledged, explained, and resolved.
In man, antibiotic exposure is associated with numerous side-effects [20] including suppression of the GMB and weakening of resistance to enteric pathobionts [21, 22]. Gut overgrowth by healthcare-associated pathogens such as Clostridium difficile and vancomycin-resistant Enterococci [11, 23, 24] imposes substantial morbidity, mortality, and economic burden [25]. Such considerations limit human antibiotic studies to clinical samples, thus constraining control over demographic (age [26], socioeconomic [27]), intrinsic (genetic [28]), and other factors (diet [29], physical activity [30], colonic transit time [31], medications [32], morbidity, environment [33]) that can affect the human GMB and/or metabolome [34]. Not surprisingly, many chemicals within the human metabolome show very high inter- and intra-individual variance [3, 35, 36]. Ingestion of a single cup of coffee, for instance, can elevate excretion of certain GMB-derived small phenolic chemicals by several-hundred-fold [37].
The lower variance of chemical levels in serum [36] relative to those in urine [35] or feces [38] should make the human serum metabolome particularly useful for studying antibiotic effects. The small number of such reports [14-18] may be due to both technical and statistical barriers. Identification and quantification of chemicals using a targeted analytic approach requires having authentic standards and generating calibration curves for each analyte of interest. This can be highly resource intensive. Untargeted approaches that draw on spectral libraries provide only semi-quantitative information and putative identification but generate data on thousands of chemicals [39]. To deal with the resulting large numbers of variables, the False Discovery Rate (FDR) [40] is commonly applied. Such a statistical constraint may be excessively strict at times, resulting in type II statistical errors. Given the bias against publishing negative results [41], investigators may be discouraged from conducting investigations of antibiotic effects on the serum metabolome or from publishing ostensibly negative data from completed studies.
A complementary approach would be to a priori identify a discrete number of chemicals expected to be consistently detectable as antibiotic-responsive by univariate analysis. Such anchor points would confirm antibiotic effects overall and justify the search for additional sensitive chemicals. Accordingly, we conducted untargeted metabolomic analysis of serum from patients treated with wide-spectrum antibiotic and then subjected the aggregate data to a focused, pre-planned analysis guided by results from animal studies.
The study protocol was approved by the Institutional Review Board of the Veterans Affairs Northeast Ohio Healthcare System (VA NEOHS). Newly hospitalized patients who were prescribed broad-spectrum antibiotic regimens were recruited for enrollment. Exclusion criteria included systemic antibiotic treatment within the month before admission, altered mental status or dementia, lack of availability of blood serum specimens prior to the start of antibiotic therapy, anticipated length of hospital stay less than 3 days, tube feeds, and end-stage renal or liver disease. Serum specimens were collected prior to the start of antibiotic therapy, on day 2 to 3 of therapy, and 3 to 5 days after completion of antibiotic therapy. To evaluate the potential for large shifts in the serum metabolome during hospitalization in the absence of antibiotic therapy, serum specimens were collected at the same time points for 5 control patients admitted to the hospital but not prescribed antibiotics. These control patients were not matched to the experimental patients based on clinical characteristics. The serum specimens were stored at -80°C. After completion of specimen collection, coded samples were shipped on dry ice to Metabolon© (Morrisville, NC) where they were inventoried and stored at -80°C until analyzed.
All sample preparation and analysis was conducted by Metabolon©. In brief, samples were extracted using proprietary methods and the resulting extract divided into 2 fractions, 1 each for analysis on gas chromatography/mass spectroscopy (GC/MS) and liquid chromatography (LC)/MS platforms [42]. The LC/MS portion of the platform was based on a Waters ACQUITY UPLC and a Thermo-Finnigan LTQ mass spectrometer, which consisted of an electrospray ionization source and linear ion-trap mass analyzer. The sample extract was split into 2 aliquots, dried, then reconstituted in acidic or basic LC-compatible solvents, each of which contained 11 or more injection standards at fixed concentrations. One aliquot was analyzed using acidic positive ion optimized conditions and the other using basic negative ion optimized conditions in 2 independent injections using separate dedicated columns. The samples destined for GC/MS analysis were re-dried under vacuum desiccation for a minimum of 24 hours prior to being derivatized under dried nitrogen using bistrimethyl-silyl-triflouroacetamide. The GC column was 5% phenyl and the temperature ramp was from 40° to 300° C in a 16-minute period. Samples were analyzed on a Thermo-Finnigan Trace DSQ fast-scanning single-quadrupole mass spectrometer using electron impact ionization. The instrument was tuned and calibrated for mass resolution and mass accuracy on a daily basis. The information output from the raw data files was automatically extracted. Compounds were identified by comparison to library entries of purified standards or recurrent unknown entities [42] (for details see Supplementary Information, Methods).
General: For pair-wise comparisons Welch’s t-tests and/or Wilcoxon’s rank sum tests were performed (after a repeated measures ANOVA). For classification, random forest analyses were done. Statistical analyses were performed using the program “R”.
Selection of Analytes for Focused Statistical Analysis: We conducted a PubMed search (2020/04/01) using the phrase “(mouse OR rat) AND (germ-free OR antibiotic) AND [(serum OR plasma OR blood)] AND metabolome.” We evaluated the resulting articles as well as any additional relevant publications that they referenced. Antibiotic reports were considered to be relevant if they met the following criteria, i) they used a conventional animal strain, ii) the antibiotic was administered for > 6 days, iii) a control group not treated with antibiotic was included, iv) animals were maintained on a standard lab diet, and v) blood was drawn no later than 24 hours after completion of antibiotic treatment. Our final focused list consisted of chemicals whose serum levels were reported to be affected both by GF status and by treatment with at least 1 antibiotic.
From the data provided by Metabolon©, we decided a priori to exclude those chemicals for which > 50% of baseline data were missing. Given that antibiotics often suppress GMB-derived metabolites, chemicals with > 50% missing values at the post-antibiotic time point were included. To address fluctuations that can occur for reasons other than antibiotic treatment, we also decided a priori to exclude those chemicals which showed significant changes between any of the time points, PRE-, MID-, and POST-antibiotic in the control group. Finally, chemicals that were significantly different between the control group and antibiotic group at the PRE time point were also to be excluded a priori.
The list of chemicals obtained from the PubMed search was cross-referenced with the list of chemicals we measured in patient serum subject to the a priori exclusions. Chemicals common to both lists were subjected to planned t-test comparisons with significance set at P < 0.05, uncorrected for multiple comparisons.
A total of 20 antibiotic-treated patients (mean age 70.5 years, range 50-90 years, all male) and 5 controls (mean age 75.4 years, range 64-90 years, all male) completed the study. With the exception of 1 antibiotic-treated participant who provided 2 MID blood samples and no POST sample, all other participants provided PRE, MID, and POST samples. The antibiotic treatment regimens included a broad-spectrum beta-lactam antibiotic (18 piperacillin/tazobactam, 1 ertapenem, and 1 ceftriaxone) alone in 4 patients, and in combination with additional agents in 16 patients (14 intravenous vancomycin and 2 moxifloxacin). The median duration of antibiotic treatment was 5 days (range 2-20 days).
The metabolomic analysis provided putative identification for 499 chemicals. Under general analysis, 69 chemicals in the control group and 153 chemicals in the antibiotic group showed significant uncorrected POST vs PRE differences (Supplementary Table 1), but in no case did any reach the FDR required for a significant time x treatment interaction (Supplementary Table 1, Supplementary Data). The Hierarchical Clustering and a Principal Component Analysis showed a high overlap between the groups (Supplementary Figure 2). The Random Forest Analysis showed that the control group and the antibiotic-treated groups could be separated before, during, or at the end of the study with a predictive accuracy 65%-68% (Supplementary Figure 3). However, none of the chemicals identified by the biochemical importance plot (Supplementary Figure 4) could sustain the FDR for a significant time x treatment interaction (Supplementary Data [Pathway Heat Map]).
Our PubMed search identified 8 relevant studies of antibiotic-effects on serum chemical levels in mice or rats [43-50] (Table 1). We also found 7 mouse studies [1, 51-56] that cumulatively identified 21 unique chemicals affected by GF status (Table 2). Overall, we identified 10 serum chemicals reported to be affected both by GF status and by antibiotic treatment in rodents.
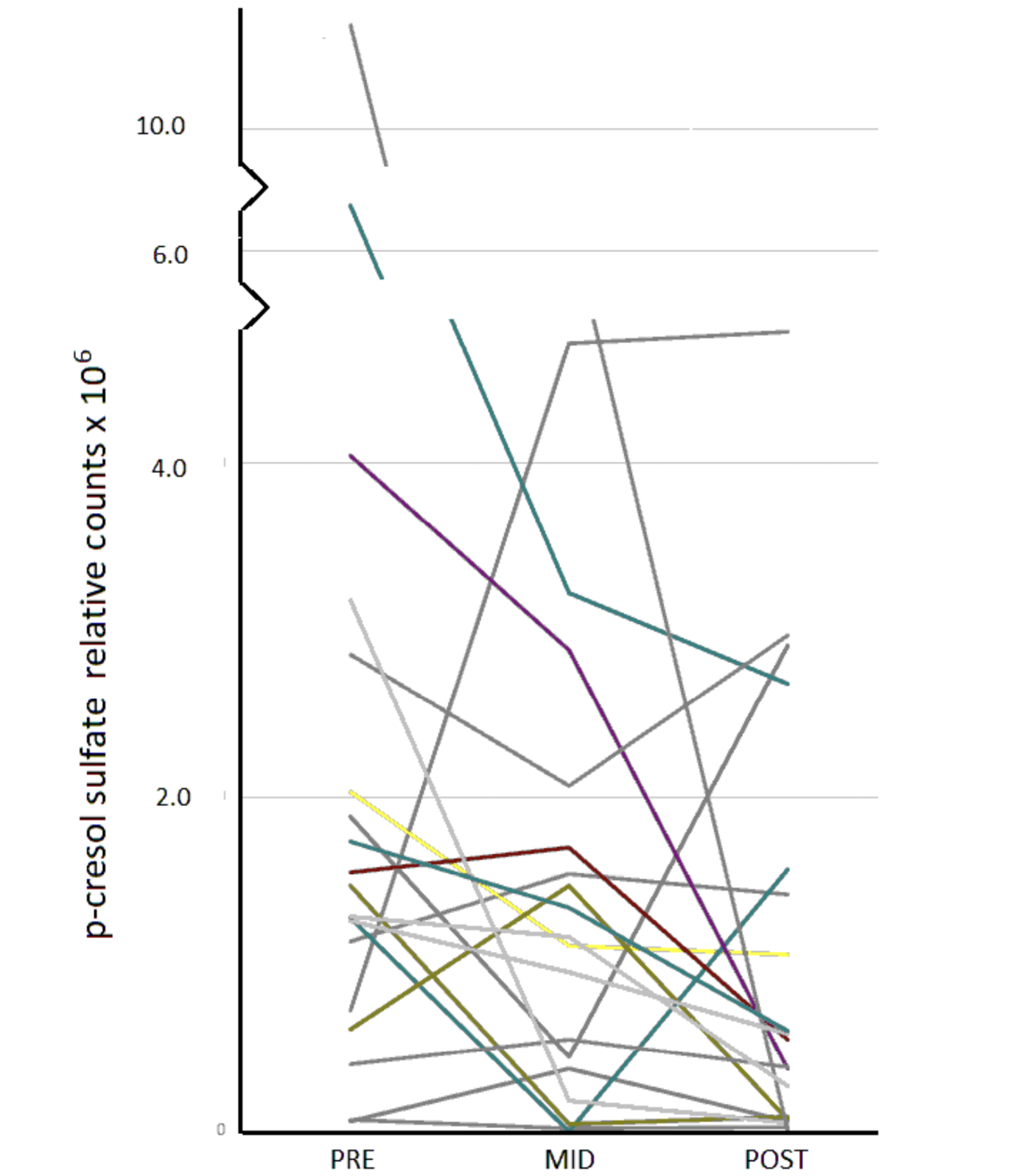
Eight such chemicals were among those measured in our human serum (Table 2, Supplementary Data [Pathway Heat Map]). N-acetyltryptophan was then excluded because > 50% of the data were missing at 1 time point. L-tyrosine was excluded because its levels at the PRE- time point tended to be higher in the antibiotic treatment group. Thus, 6 chemicals remained for consideration (Table 2). Of those, only levels of serotonin (5HT) did not show significant differences in POST vs PRE or MID vs PRE comparisons whereas levels of 5 chemicals significantly declined (Table 3). The pattern of decline over time, however, varied greatly across individuals. In the case of p-cresol sulfate, for instance, some individuals showed progressive decline over the duration of treatment, others showed maximal decline at the MID point with some recovery, and several showed a decline only after the MID point (Figure 1).
Table 1: Antibiotic Effects on Small Molecules in the Serum of the Mouse or Rat
|
|
|
|
|
|
|
[43] |
|
|
|
|
|
[44] |
|
|
|
|
|
[45] |
|
|
|
|
|
[46] |
|
|
|
|
|
[47] |
|
|
|
|
|
[48] |
|
|
|
|
|
[50] |
|
|
|
|
|
[49] |
|
|
|
|
Mouse or rat studies in which serum chemicals were reported and which satisfied the following criteria: i) conventional strain, ii) antibiotic administered for > 6 days, iii) included a non-antibiotic treated control group, iv) standard lab diet, v) blood drawn no later than 24h after the termination of antibiotic. M- male, F- female.
Table 2: Selecting Small Molecules for Focused Analysis in Serum Based on their Sensitivity to Germ-Free Status and Antibiotic Effects in Rodents
|
|
|
|
|
|
|
cinnamoylglycine |
|
|
||
|
diydroxyquinoline glucuronide |
|
|||
|
equol sulfate |
|
|||
|
hippuric acid* |
|
|
|
|
|
indole-3-propionic acid* |
|
|
|
|
|
indoxyl-3-sulfate* |
|
|
|
|
|
methyl equol sulfate |
|
|||
|
N-acetyltryptophan |
|
|
|
|
|
p-cresol sulfate* |
|
|
|
|
|
phenol sulfate* |
|
|
|
|
|
phenylacetylglycine |
|
|
||
|
phenylpropionylglycine |
|
|||
|
pyrocatechol sulfate |
|
|
||
|
serotonin* |
|
|
|
|
|
L-tryptophan |
|
|
||
|
L-tyrosine |
|
|
|
|
|
urate |
|
|
||
|
3-(3-sulfooxyphenyl) Phenyl sulfate propanoic acid |
|
|||
|
3-Carboxy-4-methyl-5-pentyl- 2-furanpropionic acid glucuronide |
|
|||
|
5-hydroxyindoleacetic acid |
|
|||
|
12-Hydroxy-5Z,8Z,10E,14Z,17Z-eicosapentaenoic acid |
|
Serum chemicals affected by germ-free status [1, 51-56], were cross-referenced with those affected by antibiotics in mouse or rat studies [43-50] and with those for which data from the current human study were available. – serum levels not detectable in GF animals, α - > 50% data missing at a relevant time point, β - trend towards significant difference between controls and antibiotic-treated group at the PRE time point, *accepted for focused analysis.
Table 3: SEffects on Serum Levels of Small Molecules: Antibiotics in Patients vs Germ-Free Status or Antibiotics in Animal Models
|
chemical |
|
|
|||||
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|||||
|
|
|
|
|||||
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|||
|
|
|
|
|
|
|
|
|
|
|
|
|
|||||
|
|
|
|
|||||
|
|
|
|
|||||
|
|
|
|
|||||
|
|
|
|
|||||
|
|
|
|
|||||
|
|
|
|
|||||
|
|
|
|
|||||
|
|
|
|
|||||
|
|
|
|
|||||
|
|
|
|
|
|
|
|
|
|
|
|
|
|||||
|
|
|
|
|||||
|
|
|
|
|||||
|
|
|
|
|||||
|
|
|
|
|
|
0.02 |
1.3 x 10-7b, 0.002a |
|
|
|
|
|
|||||
|
|
|
|
|||||
|
|
|
|
|||||
|
|
|
|
|||||
|
|
|
|
|
|
|
|
|
|
|
|
||||||
Serum ratios (POST/PRE or MID/PRE) of antibiotic (Abx)-treated patients or germ-free (GF) or Abx-treated animals/controls (CONT). BACI – bacitracin, CLIN – clindamycin, DOX – doxycycline, GENT – gentamicin, LEVO – levofloxacin, LINCO – lincomycin, MOXI – moxifloxacin, NEO – neomycin, PEN – penicillin G, POLY – polymyxin B, STREP- streptomycin, TETRA – tetracycline, VAN- vancomycin. a. Chu et al, 2019, b. Wikoff et al 2009, c. Behr et al, 2017, d. Behr et al 2018, e. Lam et al, 2016, f. Sun et al, 2013, g. Devlin et al, 2016, h. Mishima et al, 2017, i. Shimada et al, 2013, j. Sjogren et al, 2012, k. et al>, 2019, l. Cho Y et al, 2019). nd - not detected in GF animals. P-values were calculated for means. POST vs PRE or MID v PRE for human Abx, GF v CONT, Abx v CONT for animal studies. Bold values for Human Abx designate comparisons which were statistically significant p < .05. * Welch’s t-test at p < 0.2 considered significant.
In our group of patients treated with broad-spectrum antibiotics, serum levels of 5 of 6 chemicals predicted to be highly GMB-dependent by animal studies, declined significantly as measured by univariate analysis. While consistent with our hypothesis, the findings have to be critically interpreted in terms of assumptions, experimental limitations, and generalizability.
We enlisted inpatient male veterans with any of several medical conditions for which broad-spectrum antibiotic treatment was indicated. The duration of antibiotic treatment varied considerably. Our small sample of controls was not clinically matched with the treatment group. We used a commercial metabolomic service (Metabolon©) that conducted a non-targeted analysis using GC/MS or LC/MS/MS. While such an approach supports detection of a very large number of chemicals, it does so on the basis of available spectroscopic libraries rather than by direct comparisons to authentic standards. This yields presumptive identification and semi-quantitative comparisons [39]. On the other hand, the chemicals in our focused analysis have relatively simple structures and well-characterized spectral properties.
A classical ANOVA that included all the aggregate data (20 patients, 5 controls, 3 time points, 499 chemicals) did not show any time x treatment interactions that exceeded the preset q < 0.1. Part of this was due to the FDR for multiple corrections. However, even if the q value was ignored and any time x treatment interaction P < 0.05 accepted, relatively few analytes would have reached significance. This can be partially attributed to the high variance of the data as well as to the unequal sizes of the control (n = 5) and antibiotic-treated (n = 20) groups. We dealt with these issues by imposing criteria determined a priori to restrict the number of chemicals admitted to a focused univariate analysis.

Figure 1. Serum levels of p-cresol sulfate immediately before (PRE), during (MID), and within 24h of completion (POST) of a course of broad-spectrum antibiotics.
The requirement that serum levels of a given chemical show GMB-dependence in both GF and antibiotic animal studies had by far the largest overall impact. Our PubMed search identified only 10 such serum chemicals (Table 2). Of those, phenylacetylglycine and pyrocatechol sulfate were not part of the panel of chemicals measured in our human serum (Table 2). Thus, only 8, or 1.6% of the total number (n = 499) of putatively identified chemicals could be considered for a focused evaluation. We also decided a priori to exclude chemicals whose levels in the control group showed significant changes during the experimental period. It turned out, however, that this rule had no practical effect and did not further limit the chemicals identified as antibiotic-responsive in animal studies (Table 2). Second, we decided to exclude chemicals that at the PRE (ie, baseline) time point already tended to be significantly different (P < 0.1) between controls and antibiotic-treated individuals. Such differences may be attributable to chance, underlying illness, or other factors. L-tyrosine was excluded in this way. Third, we decided to exclude all chemicals for which > 50% of data were missing for any time point in the controls or at the PRE time point in the antibiotic group. This was based on the fact that GF-status [1, 52, 53] or antibiotic treatment [43-48] was much more likely to lower serum levels of a chemical than to elevate it (Table 3). Hence, failure to detect a chemical after initiation of antibiotic treatment could reflect antibiotic-induced suppression of that chemical. However, had the > 50% missing data exclusion been applied universally, it would not have affected the exclusion of N-acetyltryptophan, for which > 80% of levels were unavailable across all time points in both groups (Supplementary Data).
Of the roughly 1014 cells comprising the human organism ~ 90% are exogenous, with the vast majority contributed by the GMB [57]. Cell density in the colon exceeds that for any other known ecosystem [58]. To meet its bioenergetic needs, each gut microbe must generate energy, maintain a redox balance, and acquire carbon and nitrogen for synthesis of necessary chemicals [59]; to achieve this, it must import needed substrates and export unwanted products. Intraluminally generated chemicals which cross the gut wall become constituents of the host’s metabolome. Accordingly, attempts to define the GMB-sensitive metabolome often start with identification of chemicals that individual microbes can absorb, metabolize, synthesize and then release into the gut lumen.
Taxonomic profiling, commonly based on deep sequencing of 16s rRNA, identifies the type and number of elements of the GMB. Functional profiling supports development of integrated catalogues of the human fecal microbial metagenome that characterize the metabolic capacity of the GMB [60]. The latter, in combination with transcriptomics, proteomics, and metabolomics can define which microbial genes are actually expressed under given conditions. Some enzymatic capacities within the human organism are GMB exclusive, others can be located only within endogenous human cells, and the remainder can be expressed in both [61]. A comparison of such GMB-derived data with homologous information about the metabolic potential of endogenous human cells [62] can suggest the origin of given chemicals within the human metabolome. Confirmatory information, however, must be derived from in vivo studies.
Homologous data can be readily obtained from gnotobiotic animal models, the extreme being the GF organism, reared from birth without a detectable GMB [7]. Thus, sensitivity to GF status was the initial inclusion criterion for our focused list of chemicals in serum (Table 2, Table 3) [1, 51-56]. Of the 21 chemicals so identified, 10 were not detectable or almost undetectable in the GF state (Table 2) and hence likely to be exclusively dependent on the GMB.
The presence of a healthy GMB is critical to the development of multiple systems including internal barriers (gut-blood, blood-brain) and end organs; the GF rodent is not just a normal animal in which the GMB has suddenly been eliminated [7]. Furthermore, the GF state does not model the multiple adaptive changes that take place within a GMB exposed to antibiotics. Such primary antibiotic-induced effects on GMB architecture can secondarily affect luminal levels of chemicals absorbed or secreted by the antibiotic-adapted GMB [10, 11, 22]. For instance, when antibiotic treatment suppresses certain constituents of the GMB, hypervirulent strains of C. difficile can exploit that niche by generating concentrations of p-cresol which are bacteriostatic to other microbes [63-65]. Alternatively, one group of microbes can provide chemicals needed by a second group of microbes which in turn promotes an environment favorable to the growth of the first [66]. Furthermore, the GMB is characterized by a high degree of metabolic redundancy; most metabolic pathways are encoded within functional genetic categories distributed across multiple types of bacteria rather than being exclusive to one genus or strain [61, 67]. Thus, antibiotics are unlikely to suppress or eliminate metabolites to the extent achieved by GF status.
For these reasons, we imposed an additional constraint on our focused list of samples, namely that serum chemicals had to be affected by antibiotic treatment in rodents. Although GMB diversity in the mouse is relatively closer to that of man [68, 69] we accepted both mouse and rat studies. We also accepted studies using any combinations of male or female animals, sexual dimorphism notwithstanding [44, 53, 70]. We did not consider metabolome data derived from matrices other than serum, given that changes across these compartments (eg, urine, feces) often correlate poorly [44, 45, 71].
A PubMed Search (initial search terms: [(serum OR plasma OR blood) AND antibiotic AND metabolome AND human]) yielded only 5 heterogeneous human serum studies of antibiotic effects [14-17]. An 8-week treatment of patients (n = 20) with cirrhosis and minimal hepatic encephalopathy with rifaximin, a poorly absorbed antibiotic with minimal effect on the GMB [18], elevated serum levels of several saturated as well as unsaturated fatty acids [72]. In the only available double-blind study, overweight or obese men (n = 57) were randomly assigned to receive placebo, amoxicillin (broad-spectrum), or vancomycin (narrow-spectrum) for 7 days; a very small number of serum chemicals were measured [14]. Vancomycin treatment elevated fecal primary bile acid levels over 5-fold and lowered fecal butyrate to below 20% of pre-treatment levels, but those changes were not reflected in serum [14]. In an open study, patients with chronic obstructive pulmonary disease were randomized to continue standard treatment with a combination of inhaled β-adrenergic agonists and inhaled corticosteroids (n = 60) or to receive add on doxycycline (n = 60) for 90 days [16]. An untargeted metabolomic analysis associated doxycycline treatment with elevations of serum citrate, imidazole, and L-arginine but a lowering of lactate and an unspecified fatty acid [16]. Of note, levels of several serum metabolites also rose in the control group [16]. In a recent study, blood samples drawn early (0 – 2 days) and late (25 days +) during treatment of infective endocarditis with any of several antibiotics, showed a lowering of several amino acids (L-tyrosine, L-valine, L-leucine, L-isoleucine) and other chemicals (glucose, mannose, unspecified polyol) [17]. Overall, the heterogeneity of antibiotics, duration of treatment, and panels of evaluated chemicals in these studies does not justify generalized conclusions.
There are many steps between the appearance of a molecule in the gut lumen, whether as an endogenous or microbial product, and its entry into peripheral serum (Figure 2). Transport across cell walls and other barriers may be passive or active and affected by a number of genetic and other factors [34]. Biotransformations may take place in the intestinal lumen, in the enterocytes lining the lumen, in the liver, kidney, or other organs [34]. Hence, both transport and biotransformation of chemicals must be considered when interpreting changes in the antibiotic-affected serum metabolome. To minimize possible primary endogenous metabolic factors, we excluded patients with end-stage renal or hepatic disease. While, in general, antibiotics have larger effects on the GMB than on hepatic or renal function or on transport mechanisms [73-75], each chemical of interest should be evaluated individually.
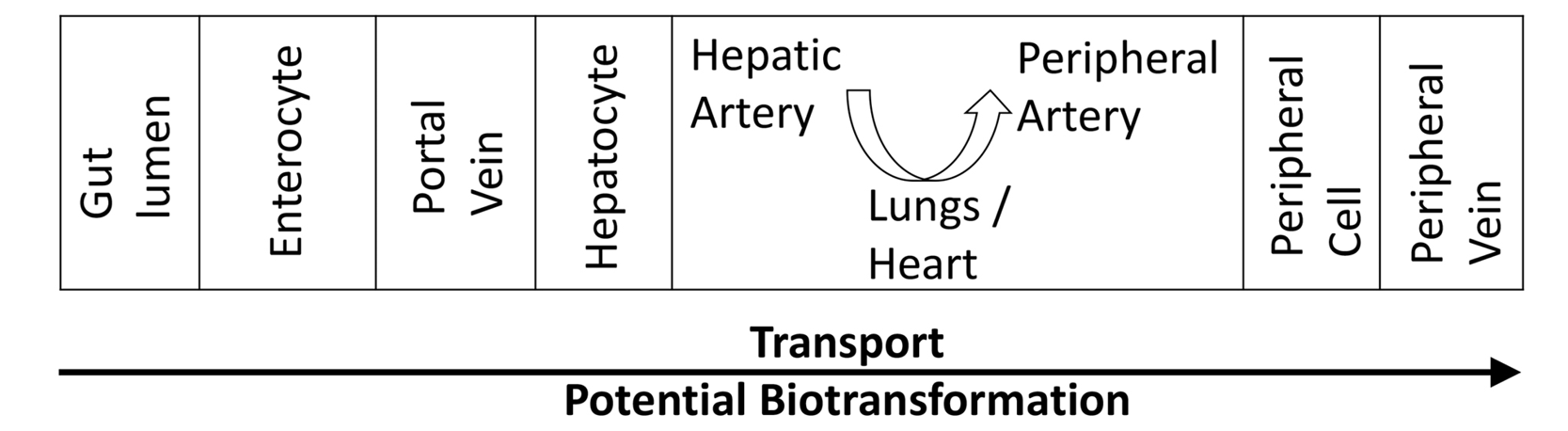
Figure 2. In order for a chemical from the gut lumen to reach a peripheral vein and become available for conventional blood sampling, it must be transported through multiple cells and compartments within which it is vulnerable to biotransformation.
The serum chemicals we identified as antibiotic-sensitive in man belong to the phenolic (hippurate, p-cresol sulfate, phenol-sulfate) or indolic (indoxyl sulfate, indole propionate) pathways. Hippuric acid is formed through the conjugation of benzoic acid (BA) with the hydrophilic moiety glycine [76, 77] in a phase II process facilitating removal of the hydrophobic but potentially toxic BA [78, 79]. Benzoic acid itself cannot be synthesized by endogenous human enzymes [62]. On the other hand mixed fecal cultures or certain constituents of the human GMB can, under anaerobic conditions, generate BA or its hydroxylated metabolites from a variety of precursors, including free L-tyrosine or L-phenylalanine [80, 81], 4-hydroxyphenylpyruvic acid [81], polyphenol rich dietary components (eg, berries, black tea, bran, fruit juices, wine) [82-89], or pure polyphenols themselves (eg, catechin, epicatechin, proanthocyanidins, quercetin) [90-93].
While HA and its hydroxylated metabolites are constituents of the human fecal metabolome [94], the GMB cannot glycinate BA to generate HA. In man, such glycination has been demonstrated in the liver and kidney [77, 95], but whether it can also take place within enterocytes, as is the case in the rat [96], is not known. In any case, the conjugation occurs via a coenzyme-A-dependent process localized to the mitochondrial matrix [97].
Similarly, neither phenol nor p-cresol can be synthesized in appreciable quantities by endogenous human enzymes [62] but can be generated under anaerobic conditions by GMB from free L-tyrosine [80, 81, 98-102] or dietary polyphenols [103]. Phenol and p-cresol are considered end-products of the GMB in that they are not further broken down in the intestinal lumen [104]. Both occur in human serum almost exclusively as sulfated conjugates (Supplementary Data) with p-cresol glucuronide a minor metabolite [105]. Aryl sulfotransferase activity capable of sulfating phenol is expressed in human feces [106] and can be suppressed by antibiotics in animal models [107]. In man, phenol-sulfotransferase activity is expressed within tissue of the small intestine [108, 109], even more so in liver, and more modestly in the kidney [109, 110]. Colonic tissue remains to be similarly evaluated. Thus, while both phenol and p-cresol are generated exclusively by the GMB, the relative importance of multiple potential loci of sulfation is not known. Antibiotic effects on their sulfation or glucuronidation are not known.
While antibiotic treatment is not known to affect conjugation of BA or p-cresol; effects on their transport must be considered separately. Gut luminal BA crosses the basilar membrane of enterocytes via monocarboxylic transporters [111, 112]. Antibiotics can markedly suppress GMB-mediated synthesis of short chain fatty acids [113], which in turn, can act both as substrates and inducers for colonocyte monocarboxylic transporters [114]; thereby potentially affecting transport of other substrates such as BA. The transport of free p-cresol across the human gut wall remains to be similarly characterized, but in general, fecal levels of free p-cresol correlate with serum levels of its conjugates [115].
Sulfated conjugates of phenol and p-cresol can be detected in human feces [115, 116]. In addition, p-cresol sulfate can be taken up by Breast Cancer Resistance Protein members of the ATP-binding cassette (ABC) protein transporter family [117] expressed on the apical membrane of human colonocytes [118, 119], as well as by organic anion transporters (OAT) (hOAT1, hOAT3) on human kidney cells [120-124]. The expression of hOAT3 in tissue of the small intestine has been detected by some investigators, but not by others [119, 125, 126], and hOAT1 and hOAT3 may also be expressed in rectal tissue [127]. Certain antibiotic (eg, penicillins, cephalosporins) can inhibit OAT-mediated uptake of substrates [128-131]. However, this would limit renal excretion of such chemicals and; hence, elevate their serum levels. Since levels of our targeted analytes were lowered by antibiotic treatment, antibiotic effects on OAT-mediated transport are unlikely.
Constituents of the human GMB readily convert L-TRP to indole [80, 104, 132-134]; endogenous human enzymes cannot do so [62]. Indole is the most abundant L-TRP metabolite of the normal human fecal metabolome (~ 3mM) [3, 135] and its levels correlate with those of serum indoxyl sulfate [115]. The conventional wisdom is that intraluminally-formed indole is transported across the colonic wall, enters the portal circulation and reaches hepatocytes, where microsomal cytochrome CYP2E1 3-hydroxylates the indole to form indoxyl [136, 137]. The latter, in turn can be sulfated to indoxyl sulfate, secreted into blood [138] and then cleared by the kidney. However, indoxyl sulfate itself is a constituent of the human fecal metabolome [115], raising the possibility that some fraction of it is either generated within the GMB or is secreted into the lumen by the host. Tracer studies in the rat, for instance, show that a small fraction of systemically administered indole enters the bile as indoxyl sulfate [139]. Furthermore, indoxyl sulfate taken up from the systemic circulation can be directly secreted into the intestinal lumen under conditions of renal failure [140]. Intriguingly, a recent rat study found no significant differences in postprandial levels of indoxyl sulfate between the portal and systemic circulations [141]. This suggests that under usual conditions, most plasma indoxyl sulfate is generated within the GMB and/or enterocytes, without a significant hepatic contribution. Comparable studies in primates have not been conducted. In any case, indoxyl sulfate is cleared mainly through renal excretion mediated at least in part by hOAT3, in a process that can be inhibited by ciprofloxacin [131]. While 2 of our patients were treated with moxifloxacin, another quinolone antibiotic, any inhibition would elevate, not lower indoxyl sulfate levels as we observed in our data.
The kinetics of indole propionic acid are less well characterized. While human cells can successively metabolize L-TRP to indole-3-pyruvic acid and then to indole-3-lactic acid [62, 142, 143], only certain constituents of the GMB are known to metabolize L-TRP through to indole propionic acid [133, 143-147]. That reductive pathway may involve indole-3-lactic acid, indol-3-acrylic acid, or other intermediates [143, 148]. Indole propionic acid can inhibit the human proton-coupled amino acid transporter in the intestinal epithelium but is not itself a transport substrate for it [149]. Thus, the transport of indole propionic acid across the gut wall remains to be characterized. In man, indole propionic acid does not undergo phase II metabolism and has been detected only as the parent compound [3]. Antibiotic effects on the transport of indole propionic acid are not known.
5HT was the only chemical whose serum levels were affected both by GF and antibiotic status in rodents (Table 2) but which was not significantly affected by antibiotic treatment in our focused analysis (Table 3). It is also the only chemical in our list that can be generated endogenously by mammalian cells [62]. Isolated reports that certain GMB constituents can synthesize 5HT directly from L-TRP [150] notwithstanding, there is no evidence that GMB-derived 5HT significantly contributes to serum levels of 5HT. On the other hand, over 90% of endogenous 5HT in the rat is synthesized by various components of the gut, namely, mucosal mast cells and myenteric neurons and, particularly, enterochromaffin cells, which express tryptophan hydroxylase 1, the rate-limiting enzyme for 5HT synthesis [151]. The gut-generated 5HT is taken up by platelets and distributed systemically [151]. Estimates from rodent models suggest that while over 80% of serum 5HT originates in cells endogenous to the gut of the host [152], about 50% of serum 5HT is subject to some regulation by the GMB [153].
Short chain fatty acids (SCFAs), generated via GMB-mediated fermentation of dietary fiber and protein, upregulate tryptophan hydroxylase 1 [154, 155]. Since SCFAs are absent from the intestinal lumen of the GF mouse [156, 157], GF status is associated with lower expression of tryptophan hydroxylase 1 in colonic tissue [56, 154]. This, given the outsized contribution of enterochromaffin cells to serum 5HT [151], is consistent with lower serum levels of 5HT in the GF mouse [1, 56]. Antibiotic effects in animal models are more difficult to explain. Treatment with vancomycin, an antibiotic that relatively selectively suppresses enterococci [158] produced a 3-fold elevation of serum 5HT in the rat [46]. However, changes in serum 5HT levels did not reach significance after treatment with combinations of antibiotics ([streptomycin + neomycin + bacitracin + polymyxin [46])], [ampicillin + neomycin + metronidazole + vancomycin [50] ])
intended to sterilize the gut. In contrast, a combination of ampicillin and neomycin lowered serum 5HT levels in the mouse by almost 50% [49]. The precise mechanisms that would explain these differences remain to be determined.
In developing our a priori criteria for selecting chemicals of interest we did not include data from patients with ileostomies. While the overwhelming preponderance of the total GMB reside in the colon, the bacterial density in the terminal ileum actually exceeds that of the large intestine [159]. Furthermore, a sparse microbiota can be found throughout the normal digestive tract [160, 161]. Naso-ileal tube aspirates from patients with intact colons reflect a similar GMB composition to that in ileal fluid from patients with ileostomies [162]. The enzymatic activity within ileal fluid is capable of metabolizing the flavonoid precursors of multiple phenolic metabolites [163]. At least some members of the ileal GMB express functionally significant tyrosine decarboxylase activity [164]. This suggests that the functional capacity of the remaining GMB in patients with ileostomies could, in theory, contribute to the human serum metabolome. In view of other data, however, that possibility seems remote. In patients with renal failure, colectomy is associated with either negligible (p-cresol sulfate, indoxyl sulfate) or significantly lower (HA) serum levels [165] of 3 of the metabolites identified as unequivocally GMB-dependent in animal studies (Table 2). Comparable data on serum or urine phenol sulfate or indole propionic acid levels after colectomy are not available. Not surprisingly, all the chemicals identified in our focused list are either known or suspected to be uremic solutes [115, 166].
Without any direct measures of the GMB, we cannot empirically evaluate possible associations between antibiotic-induced changes in the serum metabolome and concomitant changes in individual operational taxonomic units (OTUs) of the GMB. The available literature supports several generalizations. Most antibiotic-treatment regimens lower the overall diversity of the GMB [14, 167-170]. Furthermore, antibiotic class-specific effects on the GMB architecture have been well demonstrated [8-11]. Our antibiotic regimens included β-lactam agents, which are known to be excreted into the intestinal tract where they can dramatically alter the GMB [9]. The most common beta-lactam antibiotic treatment was the piperacillin/tazobactam combination, which provides broad-spectrum activity against anaerobes, facultative gram-negative bacilli, and enterococci [9, 10].
Most metabolic pathways are encoded within functional genetic categories distributed across multiple types of bacteria rather than being exclusive to one genus or strain [61, 67, 171]. Suppression of one taxa can create a niche opportunity for other taxa [11, 158, 172]. Furthermore, some enzymatic capacities within the human organism are GMB exclusive, others can be located only within endogenous human cells, and the remainder can be expressed in both [61]. A given GMB-derived chemical in the human metabolome does not per se carry the fingerprint of the particular microbe that generated it. Hence, antibiotic-induced lowering of serum chemicals is more likely to be detected for those chemicals generated relatively exclusively by taxa targeted by the antibiotic regimen.
All of the 5 chemicals determined to be antibiotic-responsive in our study are themselves either fermentation products or metabolites of fermentation products of aromatic amino acids. Although aromatic amino acids can be metabolized to some degree by cultures of a range of intestinal anaerobes, including members of the genera Bacteroides, Lactobacillus, Clostridia, and Bifidobacteria [173], only limited members of those genera can generate BA, p-cresol, or phenol from L-TYR and/or L-PHE [80, 98-102]. There is further specificity within strains. Clostridium difficile can ferment L-TYR, L-PHE, or L-TRP whereas C. perfringens can do so only to L-TYR [99, 144]. Furthermore, C. difficile is the only Clostridial species expressing 4-hydroxyphenylacetate decarboxylase (EC 4.1.1.83), an enzyme critical to the metabolism of L-TYR through to the end product p-cresol [63, 174].
A relatively large number of anaerobic constituents of the healthy human GMB, including E. coli, express tryptophanase (EC 4.1.99.1), which converts L-TRP to indole [80, 99, 104, 132-134]. Clostridium difficile cannot itself generate indole but can modify the intestinal milieu to promote indole-generation by other bacteria [175]. A more restricted number of microbes, including Clostridial strains, can ferment L-TRP to indole propionic acid [99, 133, 143-147].
We noted earlier, that many small phenolic molecules can also be generated by GMB-mediated degradation of polyphenol rich dietary components [82-89]. This applies to all 3 phenolic metabolites in our study (phenol sulfate, p-cresol sulfate, HA). We cannot distinguish the fraction of such GMB-generated metabolites derived from dietary polyphenols vs free L-TYR or L-PHE. The critical first step in polyphenol metabolism is cleavage of the C ring of the heterocyclic flavan nucleus, a reaction outside the capacity of endogenous human enzymes but one that can be carried out by a limited number of members of the human GMB [34], including several Clostridial species [176-179]. Regulation of the full cascade of polyphenol metabolism, however, is incompletely understood. For example, while plasma HA levels rise rapidly after the ingestion of black tea polyphenols [180], dietary polyphenols either lower or do not affect the excretion of phenol sulfate [181, 182]. A switch from a conventional Western diet to an uncooked but fiber- and polyphenol-rich vegan diet with comparable total protein content lowers both serum phenol sulfate and p-cresol sulfate [183]. This underscores that dietary constituents other than polyphenols can also affect serum levels of phenolic end products. Whether the same members of the GMB mediate all such processes remains to be determined.
Of course, data from in vitro monoculture experiments must be complemented by in vivo studies that allow for polymicrobial population dynamics. Such efforts remain highly preliminary. In a subsample (n = 855) of individuals with minimal renal decline, serum indoxyl sulfate and p-cresol sulfate were both associated with the presence of family members of Clostridiales in the GMB [184]. More recently, in a study of mixed inpatients treated clinically with antibiotics, urinary indoxyl levels were found to be associated with the relative abundance of Clostridiales [168]. Thus, our own data would suggest that the broad-spectrum antibiotic suppressed at the very least, members of the Clostridia family.
Using a clinical sample of patients and a limited number of targets we demonstrated that broad-spectrum antibiotic treatment lowered serum levels of 5 chemicals (phenol sulfate, p-cresol sulfate, HA, indoxyl sulfate, indole propionic acid) previously shown to be absent or markedly suppressed in the serum of GF mice (Table 3). Based on a consideration of transport and other mechanisms, we conclude that the most likely explanation for the data is an antibiotic-induced suppression of Clostridia and other species. All 5 chemicals, often referred to as uremic solutes were previously considered to be GMB-dependent based on various animal and human studies [115, 166]. However, ours is the first study to demonstrate that clinical administration of broad-spectrum antibiotic regimens, even for a variable period of time, significantly lowers serum levels of this group of chemicals. That is all the more remarkable given the manifold other factors known to affect GMB composition and which were not controlled in our study; these include age [26], socioeconomic status [27], genetics [28], concomitant medications [32], and particularly diet [29] and colonic transit time [31]. It is of note that in control populations, excretion of p-cresol sulfate is positively associated while phenol sulfate is negatively associated with colonic transit time [31]. The fact that serum levels of both these chemicals were lowered in our study would indicate that the effects of antibiotic-induced suppression of the GMB were larger than those of other factors which we did not control for.
In view of the increasing use of antibiotics worldwide [19], metabolomic and microbiomic investigations of such treatments remain grossly underutilized. Establishing core chemicals or profiles of such chemicals that confirm antibiotic effects should permit identification of other small molecules that respond more subtly to antibiotic-induced shifts in the GMB. Development of restricted lists of chemicals to be examined a priori could allow detection of effects that would not be apparent if statistical adjustments for comparing a very large number of variables were applied. Rapidly emerging data implicating the microbiome and its dependent metabolome across medical disciplines, including immunology [185] and neuropsychiatry [186], oblige investigators to judiciously exploit available patient samples in the service of developing novel biomarkers and treatments.
This work was supported by the Research Service of the VA Northeast Ohio Healthcare System and by a grant from the VISN 10 Research Initiative Program (to CJD and GEJ). The manuscript content is solely the responsibility of the authors and does not necessarily represent the official views of the Department of Veterans Affairs and/or the authors’ employers and/or their academically affiliated institutions.
Supplementary materials are available at the Pathogens and Immunity website. Supplementary data may be provided by the authors to benefit the reader. Supplementary data are not copyedited and are the sole responsibility of the authors. Questions or comments related to supplementary materials should be addressed to the corresponding author.
Supplementary Information, Methods
Curtis J. Donskey has received research funding from Clorox, Pfizer, and PDI. He also serves as an associate editor for Pathogens and Immunity. All other authors report no relevant conflicts of interest.
1. Wikoff WR, Anfora AT, Liu J, Schultz PG, Lesley SA, Peters EC, Siuzdak G. Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proc Natl Acad Sci U S A. 2009;106(10):3698-703. Epub 2009/02/24. doi: 10.1073/pnas.0812874106. PubMed PMID: 19234110; PMCID: PMC2656143.
2. Collins SM, Surette M, Bercik P. The interplay between the intestinal microbiota and the brain. Nat Rev Microbiol. 2012;10(11):735-42. Epub 2012/09/25. doi: 10.1038/nrmicro2876. PubMed PMID: 23000955.
3. Wishart DS, Feunang YD, Marcu A, Guo AC, Liang K, Vazquez-Fresno R, Sajed T, Johnson D, Li C, Karu N, Sayeeda Z, Lo E, Assempour N, Berjanskii M, Singhal S, Arndt D, Liang Y, Badran H, Grant J, Serra-Cayuela A, Liu Y, Mandal R, Neveu V, Pon A, Knox C, Wilson M, Manach C, Scalbert A. HMDB 4.0: the human metabolome database for 2018. Nucleic Acids Res. 2018;46(D1):D608-D17. Epub 2017/11/16. doi: 10.1093/nar/gkx1089. PubMed PMID: 29140435; PMCID: PMC5753273.
4. Brown JM, Hazen SL. Targeting of microbe-derived metabolites to improve human health: The next frontier for drug discovery. J Biol Chem. 2017;292(21):8560-8. Epub 2017/04/09. doi: 10.1074/jbc.R116.765388. PubMed PMID: 28389555; PMCID: PMC5448085.
5. Donia MS, Fischbach MA. Small molecules from the human microbiota. Science. 2015;349(6246):1254766-1-10. Epub 2015/07/25. doi: 10.1126/science.1254766. PubMed PMID: 26206939; PMCID: PMC4641445.
6. Tomas-Barberan FA, Selma MV, Espin JC. Polyphenols’ Gut Microbiota Metabolites: Bioactives or Biomarkers? J Agric Food Chem. 2018;66(14):3593-4. Epub 2018/03/21. doi: 10.1021/acs.jafc.8b00827. PubMed PMID: 29558130.
7. Luczynski P, McVey Neufeld KA, Oriach CS, Clarke G, Dinan TG, Cryan JF. Growing up in a Bubble: Using Germ-Free Animals to Assess the Influence of the Gut Microbiota on Brain and Behavior. Int J Neuropsychopharmacol. 2016;19(8). Epub 2016/02/26. doi: 10.1093/ijnp/pyw020. PubMed PMID: 26912607; PMCID: PMC5006193.
8. Dudek-Wicher RK, Junka A, Bartoszewicz M. The influence of antibiotics and dietary components on gut microbiota. Prz Gastroenterol. 2018;13(2):85-92. Epub 2018/07/14. doi: 10.5114/pg.2018.76005. PubMed PMID: 30002765; PMCID: PMC6040098.
9. Donskey CJ, Chowdhry TK, Hecker MT, Hoyen CK, Hanrahan JA, Hujer AM, Hutton-Thomas RA, Whalen CC, Bonomo RA, Rice LB. Effect of antibiotic therapy on the density of vancomycin-resistant enterococci in the stool of colonized patients. N Engl J Med. 2000;343(26):1925-32. Epub 2001/01/03. doi: 10.1056/NEJM200012283432604. PubMed PMID: 11136263; PMCID: PMC4370337.
10. Jump RL, Kraft D, Hurless K, Polinkovsky A, Donskey CJ. Impact of Tigecycline Versus Other Antibiotics on the Fecal Metabolome and on Colonization Resistance to Clostridium Difficile in Mice. Pathog Immun. 2017;2(1):1-20. Epub 2017/02/22. doi: 10.20411/pai.v2i1.159. PubMed PMID: 28217763; PMCID: PMC5315001.
11. Pultz NJ, Donskey CJ. Effect of antibiotic treatment on growth of and toxin production by Clostridium difficile in the cecal contents of mice. Antimicrob Agents Chemother. 2005;49(8):3529-32. Epub 2005/07/29. doi: 10.1128/AAC.49.8.3529-3532.2005. PubMed PMID: 16048976; PMCID: PMC1196291.
12. Lee SH, An JH, Park HM, Jung BH. Investigation of endogenous metabolic changes in the urine of pseudo germ-free rats using a metabolomic approach. J Chromatogr B Analyt Technol Biomed Life Sci. 2012;887-888:8-18. Epub 2012/02/04. doi: 10.1016/j.jchromb.2011.12.030. PubMed PMID: 22300547.
13. Rogers WF, Jr., Burdick MP, Burnett GR. The effect of antibiotics on the excretion of phenolic compounds. J Lab Clin Med. 1955;45(1):87-96. Epub 1955/01/01. PubMed PMID: 13233631.
14. Reijnders D, Goossens GH, Hermes GD, Neis EP, van der Beek CM, Most J, Holst JJ, Lenaerts K, Kootte RS, Nieuwdorp M, Groen AK, Olde Damink SW, Boekschoten MV, Smidt H, Zoetendal EG, Dejong CH, Blaak EE. Effects of Gut Microbiota Manipulation by Antibiotics on Host Metabolism in Obese Humans: A Randomized Double-Blind Placebo-Controlled Trial. Cell Metab. 2016;24(1):63-74. Epub 2016/07/15. doi: 10.1016/j.cmet.2016.06.016. PubMed PMID: 27411009.
15. Nazzal L, Roberts J, Singh P, Jhawar S, Matalon A, Gao Z, Holzman R, Liebes L, Blaser MJ, Lowenstein J. Microbiome perturbation by oral vancomycin reduces plasma concentration of two gut-derived uremic solutes, indoxyl sulfate and p-cresyl sulfate, in end-stage renal disease. Nephrol Dial Transplant. 2017;32(11):1809-17. Epub 2017/04/06. doi: 10.1093/ndt/gfx029. PubMed PMID: 28379433.
16. Singh B, Jana SK, Ghosh N, Das SK, Joshi M, Bhattacharyya P, Chaudhury K. Metabolomic profiling of doxycycline treatment in chronic obstructive pulmonary disease. J Pharm Biomed Anal. 2017;132:103-8. Epub 2016/10/28. doi: 10.1016/j.jpba.2016.09.034. PubMed PMID: 27697570.
17. Klein CF, Holle SLK, Andersen MH, Pedersen A, Bundgaard H, Iversen KK, Malmendal A. In-hospital metabolite changes in infective endocarditis-a longitudinal (1)H NMR-based study. Eur J Clin Microbiol Infect Dis. 2019;38(8):1553-60. Epub 2019/05/24. doi: 10.1007/s10096-019-03586-z. PubMed PMID: 31119577.
18. Koo HL, DuPont HL. Rifaximin: a unique gastrointestinal-selective antibiotic for enteric diseases. Curr Opin Gastroenterol. 2010;26(1):17-25. Epub 2009/11/03. doi: 10.1097/MOG.0b013e328333dc8d. PubMed PMID: 19881343; PMCID: PMC4737517.
19. Klein EY, Van Boeckel TP, Martinez EM, Pant S, Gandra S, Levin SA, Goossens H, Laxminarayan R. Global increase and geographic convergence in antibiotic consumption between 2000 and 2015. Proc Natl Acad Sci U S A. 2018;115(15):E3463-e70. Epub 2018/03/28. doi: 10.1073/pnas.1717295115. PubMed PMID: 29581252; PMCID: PMC5899442.
20. CDC. Adverse Drug Events from Antibiotics 2020 [cited 2020]. Available from: https://www.cdc.gov/medicationsafety/adverse-drug-events-specific-medicines.html#anchor_1558445387.
21. Kamada N, Chen GY, Inohara N, Nunez G. Control of pathogens and pathobionts by the gut microbiota. Nat Immunol. 2013;14(7):685-90. Epub 2013/06/20. doi: 10.1038/ni.2608. PubMed PMID: 23778796; PMCID: PMC4083503.
22. Baumler AJ, Sperandio V. Interactions between the microbiota and pathogenic bacteria in the gut. Nature. 2016;535(7610):85-93. Epub 2016/07/08. doi: 10.1038/nature18849. PubMed PMID: 27383983; PMCID: PMC5114849.
23. Pultz NJ, Stiefel U, Subramanyan S, Helfand MS, Donskey CJ. Mechanisms by which anaerobic microbiota inhibit the establishment in mice of intestinal colonization by vancomycin-resistant Enterococcus. J Infect Dis. 2005;191(6):949-56. Epub 2005/02/18. doi: 10.1086/428090. PubMed PMID: 15717271.
24. Jump RL, Polinkovsky A, Hurless K, Sitzlar B, Eckart K, Tomas M, Deshpande A, Nerandzic MM, Donskey CJ. Metabolomics analysis identifies intestinal microbiota-derived biomarkers of colonization resistance in clindamycin-treated mice. PLoS One. 2014;9(7):e101267. Epub 2014/07/06. doi: 10.1371/journal.pone.0101267. PubMed PMID: 24988418; PMCID: PMC4079339.
25. Deshpande A, Pant C, Olyaee M, Donskey CJ. Hospital readmissions related to Clostridium difficile infection in the United States. Am J Infect Control. 2018;46(3):346-7. Epub 2017/10/21. doi: 10.1016/j.ajic.2017.08.043. PubMed PMID: 29050906.
26. Fischer N, Relman DA. Clostridium difficile, Aging, and the Gut: Can Microbiome Rejuvenation Keep Us Young and Healthy? J Infect Dis. 2018;217(2):174-6. Epub 2017/10/03. doi: 10.1093/infdis/jix417. PubMed PMID: 28968708; PMCID: PMC5853914.
27. Bowyer RCE, Jackson MA, Le Roy CI, Ni Lochlainn M, Spector TD, Dowd JB, Steves CJ. Socioeconomic Status and the Gut Microbiome: A TwinsUK Cohort Study. Microorganisms. 2019;7(1). Epub 2019/01/16. doi: 10.3390/microorganisms7010017. PubMed PMID: 30641975; PMCID: PMC6351927.
28. Xie H, Guo R, Zhong H, Feng Q, Lan Z, Qin B, Ward KJ, Jackson MA, Xia Y, Chen X, Chen B, Xia H, Xu C, Li F, Xu X, Al-Aama JY, Yang H, Wang J, Kristiansen K, Wang J, Steves CJ, Bell JT, Li J, Spector TD, Jia H. Shotgun Metagenomics of 250 Adult Twins Reveals Genetic and Environmental Impacts on the Gut Microbiome. Cell Syst. 2016;3(6):572-84 e3. Epub 2016/11/08. doi: 10.1016/j.cels.2016.10.004. PubMed PMID: 27818083; PMCID: PMC6309625.
29. Singh RK, Chang HW, Yan D, Lee KM, Ucmak D, Wong K, Abrouk M, Farahnik B, Nakamura M, Zhu TH, Bhutani T, Liao W. Influence of diet on the gut microbiome and implications for human health. J Transl Med. 2017;15(1):73. Epub 2017/04/09. doi: 10.1186/s12967-017-1175-y. PubMed PMID: 28388917; PMCID: PMC5385025.
30. Allen JM, Mailing LJ, Niemiro GM, Moore R, Cook MD, White BA, Holscher HD, Woods JA. Exercise Alters Gut Microbiota Composition and Function in Lean and Obese Humans. Med Sci Sports Exerc. 2018;50(4):747-57. Epub 2017/11/23. doi: 10.1249/MSS.0000000000001495. PubMed PMID: 29166320.
31. Roager HM, Hansen LB, Bahl MI, Frandsen HL, Carvalho V, Gobel RJ, Dalgaard MD, Plichta DR, Sparholt MH, Vestergaard H, Hansen T, Sicheritz-Ponten T, Nielsen HB, Pedersen O, Lauritzen L, Kristensen M, Gupta R, Licht TR. Colonic transit time is related to bacterial metabolism and mucosal turnover in the gut. Nature microbiology. 2016;1(9):16093. Epub 2016/08/27. doi: 10.1038/nmicrobiol.2016.93. PubMed PMID: 27562254.
32. Maier L, Pruteanu M, Kuhn M, Zeller G, Telzerow A, Anderson EE, Brochado AR, Fernandez KC, Dose H, Mori H, Patil KR, Bork P, Typas A. Extensive impact of non-antibiotic drugs on human gut bacteria. Nature. 2018;555(7698):623-8. Epub 2018/03/21. doi: 10.1038/nature25979. PubMed PMID: 29555994; PMCID: PMC6108420.
33. Rothschild D, Weissbrod O, Barkan E, Kurilshikov A, Korem T, Zeevi D, Costea PI, Godneva A, Kalka IN, Bar N, Shilo S, Lador D, Vila AV, Zmora N, Pevsner-Fischer M, Israeli D, Kosower N, Malka G, Wolf BC, Avnit-Sagi T, Lotan-Pompan M, Weinberger A, Halpern Z, Carmi S, Fu J, Wijmenga C, Zhernakova A, Elinav E, Segal E. Environment dominates over host genetics in shaping human gut microbiota. Nature. 2018;555(7695):210-5. Epub 2018/03/01. doi: 10.1038/nature25973. PubMed PMID: 29489753.
34. Williamson G, Clifford MN. Role of the small intestine, colon and microbiota in determining the metabolic fate of polyphenols. Biochem Pharmacol. 2017;139:24-39. Epub 2017/03/23. doi: 10.1016/j.bcp.2017.03.012. PubMed PMID: 28322745.
35. Bouatra S, Aziat F, Mandal R, Guo AC, Wilson MR, Knox C, Bjorndahl TC, Krishnamurthy R, Saleem F, Liu P, Dame ZT, Poelzer J, Huynh J, Yallou FS, Psychogios N, Dong E, Bogumil R, Roehring C, Wishart DS. The human urine metabolome. PLoS One. 2013;8(9):e73076. Epub 2013/09/12. doi: 10.1371/journal.pone.0073076. PubMed PMID: 24023812; PMCID: PMC3762851.
36. Psychogios N, Hau DD, Peng J, Guo AC, Mandal R, Bouatra S, Sinelnikov I, Krishnamurthy R, Eisner R, Gautam B, Young N, Xia J, Knox C, Dong E, Huang P, Hollander Z, Pedersen TL, Smith SR, Bamforth F, Greiner R, McManus B, Newman JW, Goodfriend T, Wishart DS. The human serum metabolome. PLoS One. 2011;6(2):e16957. Epub 2011/03/02. doi: 10.1371/journal.pone.0016957. PubMed PMID: 21359215; PMCID: PMC3040193.
37. Stalmach A, Mullen W, Barron D, Uchida K, Yokota T, Cavin C, Steiling H, Williamson G, Crozier A. Metabolite profiling of hydroxycinnamate derivatives in plasma and urine after the ingestion of coffee by humans: identification of biomarkers of coffee consumption. Drug Metab Dispos. 2009;37(8):1749-58. Epub 2009/05/23. doi: 10.1124/dmd.109.028019. PubMed PMID: 19460943.
38. Karu N, Deng L, Slae M, Guo AC, Sajed T, Huynh H, Wine E, Wishart DS. A review on human fecal metabolomics: Methods, applications and the human fecal metabolome database. Anal Chim Acta. 2018;1030:1-24. Epub 2018/07/24. doi: 10.1016/j.aca.2018.05.031. PubMed PMID: 30032758.
39. Marshall DD, Powers R. Beyond the paradigm: Combining mass spectrometry and nuclear magnetic resonance for metabolomics. Progress in nuclear magnetic resonance spectroscopy. 2017;100:1-16. Epub 2017/05/30. doi: 10.1016/j.pnmrs.2017.01.001. PubMed PMID: 28552170; PMCID: PMC5448308.
40. Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci U S A. 2003;100(16):9440-5. Epub 2003/07/29. doi: 10.1073/pnas.1530509100. PubMed PMID: 12883005; PMCID: PMC170937.
41. Dwan K, Gamble C, Williamson PR, Kirkham JJ. Systematic review of the empirical evidence of study publication bias and outcome reporting bias - an updated review. PLoS One. 2013;8(7):e66844. Epub 2013/07/19. doi: 10.1371/journal.pone.0066844. PubMed PMID: 23861749; PMCID: PMC3702538.
42. Evans AM, Bridgewater BR, Liu Q, Mitchell MW, Robinson RJ, Dai H, Stewart SJ, DeHaven CD, Miller LAD. High resolution mass spectrometry improves data quantity and quality as compared to unit mass resolution mass spectrometry in high-throughput profiling metabolomics. Metabolomics. 2014;4(2). doi: 10.4172/2153-0769.1000132.
43. Behr C, Kamp H, Fabian E, Krennrich G, Mellert W, Peter E, Strauss V, Walk T, Rietjens I, van Ravenzwaay B. Gut microbiome-related metabolic changes in plasma of antibiotic-treated rats. Arch Toxicol. 2017;91(10):3439-54. Epub 2017/03/25. doi: 10.1007/s00204-017-1949-2. PubMed PMID: 28337503.
44. Behr C, Ramirez-Hincapie S, Cameron HJ, Strauss V, Walk T, Herold M, Beekmann K, Rietjens I, van Ravenzwaay B. Impact of lincosamides antibiotics on the composition of the rat gut microbiota and the metabolite profile of plasma and feces. Toxicol Lett. 2018;296:139-51. Epub 2018/08/14. doi: 10.1016/j.toxlet.2018.08.002. PubMed PMID: 30102961.
45. Jiang L, Zhao X, Huang C, Lei H, Tang H, Wang Y. Dynamic changes in metabolic profiles of rats subchronically exposed to mequindox. Mol Biosyst. 2014;10(11):2914-22. Epub 2014/08/19. doi: 10.1039/c4mb00218k. PubMed PMID: 25131851.
46. Lam V, Su J, Hsu A, Gross GJ, Salzman NH, Baker JE. Intestinal Microbial Metabolites Are Linked to Severity of Myocardial Infarction in Rats. PLoS One. 2016;11(8):e0160840. Epub 2016/08/10. doi: 10.1371/journal.pone.0160840. PubMed PMID: 27505423; PMCID: PMC4978455.
47. Lin W, Wang W, Yang H, Wang D, Ling W. Influence of Intestinal Microbiota on the Catabolism of Flavonoids in Mice. J Food Sci. 2016;81(12):H3026-H34. Epub 2016/10/30. doi: 10.1111/1750-3841.13544. PubMed PMID: 27792839.
48. Sun J, Schnackenberg LK, Khare S, Yang X, Greenhaw J, Salminen W, Mendrick DL, Beger RD. Evaluating effects of penicillin treatment on the metabolome of rats. J Chromatogr B Analyt Technol Biomed Life Sci. 2013;932:134-43. Epub 2013/07/09. doi: 10.1016/j.jchromb.2013.05.030. PubMed PMID: 23831706.
49. Martin AM, Yabut JM, Choo JM, Page AJ, Sun EW, Jessup CF, Wesselingh SL, Khan WI, Rogers GB, Steinberg GR, Keating DJ. The gut microbiome regulates host glucose homeostasis via peripheral serotonin. Proc Natl Acad Sci U S A. 2019;116(40):19802-4. Epub 2019/09/19. doi: 10.1073/pnas.1909311116. PubMed PMID: 31527237; PMCID: PMC6778212.
50. Cho Y, Osgood RS, Bell LN, Karoly ED, Shore SA. Ozone-induced changes in the serum metabolome: Role of the microbiome. PLoS One. 2019;14(8):e0221633. Epub 2019/08/28. doi: 10.1371/journal.pone.0221633. PubMed PMID: 31454377; PMCID: PMC6711505
51. Devlin AS, Marcobal A, Dodd D, Nayfach S, Plummer N, Meyer T, Pollard KS, Sonnenburg JL, Fischbach MA. Modulation of a circulating uremic solute via rational genetic manipulation of the gut microbiota. Cell Host Microbe. 2016;20(6):709-15. Epub 2016/12/06. doi: 10.1016/j.chom.2016.10.021. PubMed PMID: 27916477; PMCID: PMC5159218.
52. Chu C, Murdock MH, Jing D, Won TH, Chung H, Kressel AM, Tsaava T, Addorisio ME, Putzel GG, Zhou L, Bessman NJ, Yang R, Moriyama S, Parkhurst CN, Li A, Meyer HC, Teng F, Chavan SS, Tracey KJ, Regev A, Schroeder FC, Lee FS, Liston C, Artis D. The microbiota regulate neuronal function and fear extinction learning. Nature. 2019;574(7779):543-8. Epub 2019/10/28. doi: 10.1038/s41586-019-1644-y. PubMed PMID: 31645720; PMCID: PMC6818753.
53. Clarke G, Grenham S, Scully P, Fitzgerald P, Moloney RD, Shanahan F, Dinan TG, Cryan JF. The microbiome-gut-brain axis during early life regulates the hippocampal serotonergic system in a sex-dependent manner. Mol Psychiatry. 2013;18(6):666-73. Epub 2012/06/13. doi: 10.1038/mp.2012.77. PubMed PMID: 22688187.
54. Mishima E, Fukuda S, Mukawa C, Yuri A, Kanemitsu Y, Matsumoto Y, Akiyama Y, Fukuda NN, Tsukamoto H, Asaji K, Shima H, Kikuchi K, Suzuki C, Suzuki T, Tomioka Y, Soga T, Ito S, Abe T. Evaluation of the impact of gut microbiota on uremic solute accumulation by a CE-TOFMS-based metabolomics approach. Kidney Int. 2017;92(3):634-45. Epub 2017/04/12. doi: 10.1016/j.kint.2017.02.011. PubMed PMID: 28396122.
55. Shimada Y, Kinoshita M, Harada K, Mizutani M, Masahata K, Kayama H, Takeda K. Commensal bacteria-dependent indole production enhances epithelial barrier function in the colon. PLoS One. 2013;8(11):e80604. Epub 2013/11/28. doi: 10.1371/journal.pone.0080604. PubMed PMID: 24278294; PMCID: PMC3835565.
56. Sjogren K, Engdahl C, Henning P, Lerner UH, Tremaroli V, Lagerquist MK, Backhed F, Ohlsson C. The gut microbiota regulates bone mass in mice. J Bone Miner Res. 2012;27(6):1357-67. Epub 2012/03/13. doi: 10.1002/jbmr.1588. PubMed PMID: 22407806; PMCID: PMC3415623.
57. Savage DC. Microbial ecology of the gastrointestinal tract. Annu Rev Microbiol. 1977;31:107-33. Epub 1977/01/01. doi: 10.1146/annurev.mi.31.100177.000543. PubMed PMID: 334036.
58. Whitman WB, Coleman DC, Wiebe WJ. Prokaryotes: the unseen majority. Proc Natl Acad Sci U S A. 1998;95(12):6578-83. Epub 1998/06/17. doi: 10.1073/pnas.95.12.6578. PubMed PMID: 9618454; PMCID: PMC33863.
59. Fischbach MA, Sonnenburg JL. Eating for two: how metabolism establishes interspecies interactions in the gut. Cell Host Microbe. 2011;10(4):336-47. Epub 2011/10/25. doi: 10.1016/j.chom.2011.10.002. PubMed PMID: 22018234; PMCID: PMC3225337.
60. Li J, Jia H, Cai X, Zhong H, Feng Q, Sunagawa S, Arumugam M, Kultima JR, Prifti E, Nielsen T, Juncker AS, Manichanh C, Chen B, Zhang W, Levenez F, Wang J, Xu X, Xiao L, Liang S, Zhang D, Zhang Z, Chen W, Zhao H, Al-Aama JY, Edris S, Yang H, Wang J, Hansen T, Nielsen HB, Brunak S, Kristiansen K, Guarner F, Pedersen O, Dore J, Ehrlich SD, Meta HITC; Bork P, Wang J, Meta HITC. An integrated catalog of reference genes in the human gut microbiome. Nat Biotechnol. 2014;32(8):834-41. Epub 2014/07/07. doi: 10.1038/nbt.2942. PubMed PMID: 24997786.
61. Sridharan GV, Choi K, Klemashevich C, Wu C, Prabakaran D, Pan LB, Steinmeyer S, Mueller C, Yousofshahi M, Alaniz RC, Lee K, Jayaraman A. Prediction and quantification of bioactive microbiota metabolites in the mouse gut. Nature communications. 2014;5:5492. Epub 2014/11/21. doi: 10.1038/ncomms6492. PubMed PMID: 25411059.
62. Kanehisa M, Furumichi M, Tanabe M, Sato Y, Morishima K. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017;45(D1):D353-D61. Epub 2016/12/03. doi: 10.1093/nar/gkw1092. PubMed PMID: 27899662; PMCID: PMC5210567.
63. Selmer T, Andrei PI. p-Hydroxyphenylacetate decarboxylase from Clostridium difficile. A novel glycyl radical enzyme catalysing the formation of p-cresol. Eur J Biochem. 2001;268(5):1363-72. Epub 2001/03/07. doi: 10.1046/j.1432-1327.2001.02001.x. PubMed PMID: 11231288.
64. Dawson LF, Donahue EH, Cartman ST, Barton RH, Bundy J, McNerney R, Minton NP, Wren BW. The analysis of para-cresol production and tolerance in Clostridium difficile 027 and 012 strains. BMC Microbiol. 2011;11:86. Epub 2011/04/30. doi: 10.1186/1471-2180-11-86. PubMed PMID: 21527013; PMCID: PMC3102038.
65. Passmore IJ, Letertre MPM, Preston MD, Bianconi I, Harrison MA, Nasher F, Kaur H, Hong HA, Baines SD, Cutting SM, Swann JR, Wren BW, Dawson LF. Para-cresol production by Clostridium difficile affects microbial diversity and membrane integrity of Gram-negative bacteria. PLoS Pathog. 2018;14(9):e1007191. Epub 2018/09/13. doi: 10.1371/journal.ppat.1007191. PubMed PMID: 30208103; PMCID: PMC6135563.
66. Rakoff-Nahoum S, Foster KR, Comstock LE. The evolution of cooperation within the gut microbiota. Nature. 2016;533(7602):255-9. Epub 2016/04/26. doi: 10.1038/nature17626. PubMed PMID: 27111508; PMCID: PMC4978124.
67. Turnbaugh PJ, Ley RE, Hamady M, Fraser-Liggett CM, Knight R, Gordon JI. The human microbiome project. Nature. 2007;449(7164):804-10. Epub 2007/10/19. doi: 10.1038/nature06244. PubMed PMID: 17943116; PMCID: PMC3709439.
68. Nagpal R, Wang S, Solberg Woods LC, Seshie O, Chung ST, Shively CA, Register TC, Craft S, McClain DA, Yadav H. Comparative Microbiome Signatures and Short-Chain Fatty Acids in Mouse, Rat, Non-human Primate, and Human Feces. Front Microbiol. 2018;9:2897. Epub 2018/12/18. doi: 10.3389/fmicb.2018.02897. PubMed PMID: 30555441; PMCID: PMC6283898.
69. Li D, Chen H, Mao B, Yang Q, Zhao J, Gu Z, Zhang H, Chen YQ, Chen W. Microbial Biogeography and Core Microbiota of the Rat Digestive Tract. Sci Rep. 2017;8:45840. Epub 2017/04/05. doi: 10.1038/srep45840. PubMed PMID: 28374781; PMCID: PMC5379200.
70. Harrison CA, Laubitz D, Midura-Kiela MT, Jamwal DR, Besselsen DG, Ghishan FK, Kiela PR. Sexual Dimorphism in the Response to Broad-spectrum Antibiotics During T Cell-mediated Colitis. J Crohns Colitis. 2019;13(1):115-26. Epub 2018/09/27. doi: 10.1093/ecco-jcc/jjy144. PubMed PMID: 30252029; PMCID: PMC6302957.
71. Liu Z, Xia B, Saric J, Utzinger J, Holmes E, Keiser J, Li JV. Effects of Vancomycin and Ciprofloxacin on the NMRI Mouse Metabolism. J Proteome Res. 2018;17(10):3565-73. Epub 2018/09/06. doi: 10.1021/acs.jproteome.8b00583. PubMed PMID: 30183313.
72. Bajaj JS, Heuman DM, Sanyal AJ, Hylemon PB, Sterling RK, Stravitz RT, Fuchs M, Ridlon JM, Daita K, Monteith P, Noble NA, White MB, Fisher A, Sikaroodi M, Rangwala H, Gillevet PM. Modulation of the metabiome by rifaximin in patients with cirrhosis and minimal hepatic encephalopathy. PLoS One. 2013;8(4):e60042. Epub 2013/04/09. doi: 10.1371/journal.pone.0060042. PubMed PMID: 23565181; PMCID: PMC3615021.
73. Westphal JF, Vetter D, Brogard JM. Hepatic side-effects of antibiotics. J Antimicrob Chemother. 1994;33(3):387-401. Epub 1994/03/01. doi: 10.1093/jac/33.3.387. PubMed PMID: 8040106.
74. Fanos V, Cataldi L. Renal transport of antibiotics and nephrotoxicity: a review. J Chemother. 2001;13(5):461-72. Epub 2002/01/05. doi: 10.1179/joc.2001.13.5.461. PubMed PMID: 11760211.
75. Barnett LMA, Cummings BS. Nephrotoxicity and Renal Pathophysiology: A Contemporary Perspective. Toxicol Sci. 2018;164(2):379-90. Epub 2018/06/26. doi: 10.1093/toxsci/kfy159. PubMed PMID: 29939355.
76. Lees HJ, Swann JR, Wilson ID, Nicholson JK, Holmes E. Hippurate: the natural history of a mammalian-microbial cometabolite. J Proteome Res. 2013;12(4):1527-46. Epub 2013/01/25. doi: 10.1021/pr300900b. PubMed PMID: 23342949.
77. Quick AJ. The conjugation of benzoic acid in man. J Biol Chem. 1931;93.
78. Iyanagi T. Molecular mechanism of phase I and phase II drug-metabolizing enzymes: implications for detoxification. Int Rev Cytol. 2007;260:35-112. Epub 2007/05/08. doi: 10.1016/s0074-7696(06)60002-8. PubMed PMID: 17482904.
79. Jancova P, Anzenbacher P, Anzenbacherova E. Phase II drug metabolizing enzymes. Biomed Pap Med Fac Univ Palacky Olomouc Czech Repub. 2010;154(2):103-16. Epub 2010/07/30. doi: 10.5507/bp.2010.017. PubMed PMID: 20668491.
80. Russell WR, Duncan SH, Scobbie L, Duncan G, Cantlay L, Calder AG, Anderson SE, Flint HJ. Major phenylpropanoid-derived metabolites in the human gut can arise from microbial fermentation of protein. Mol Nutr Food Res. 2013;57(3):523-35. Epub 2013/01/26. doi: 10.1002/mnfr.201200594. PubMed PMID: 23349065.
81. Curtius HC, Mettler M, Ettlinger L. Study of the intestinal tyrosine metabolism using stable isotopes and gas chromatography-mass spectrometry. J Chromatogr. 1976;126:569-80.
82. Koistinen VM, Nordlund E, Katina K, Mattila I, Poutanen K, Hanhineva K, Aura AM. Effect of Bioprocessing on the In Vitro Colonic Microbial Metabolism of Phenolic Acids from Rye Bran Fortified Breads. J Agric Food Chem. 2017;65(9):1854-64. Epub 2017/02/17. doi: 10.1021/acs.jafc.6b05110. PubMed PMID: 28206756.
83. Vetrani C, Rivellese AA, Annuzzi G, Adiels M, Boren J, Mattila I, Oresic M, Aura AM. Metabolic transformations of dietary polyphenols: comparison between in vitro colonic and hepatic models and in vivo urinary metabolites. J Nutr Biochem. 2016;33:111-8. Epub 2016/05/09. doi: 10.1016/j.jnutbio.2016.03.007. PubMed PMID: 27155917.
84. Brown EM, McDougall GJ, Stewart D, Pereira-Caro G, Gonzalez-Barrio R, Allsopp P, Magee P, Crozier A, Rowland I, Gill CI. Persistence of anticancer activity in berry extracts after simulated gastrointestinal digestion and colonic fermentation. PLoS One. 2012;7(11):e49740. Epub 2012/11/28. doi: 10.1371/journal.pone.0049740. PubMed PMID: 23185422; PMCID: PMC3504104.
85. Brown EM, Nitecki S, Pereira-Caro G, McDougall GJ, Stewart D, Rowland I, Crozier A, Gill CI. Comparison of in vivo and in vitro digestion on polyphenol composition in lingonberries: potential impact on colonic health. Biofactors. 2014;40(6):611-23. Epub 2014/11/02. doi: 10.1002/biof.1173. PubMed PMID: 25359330.
86. Stalmach A, Edwards CA, Wightman JD, Crozier A. Colonic catabolism of dietary phenolic and polyphenolic compounds from Concord grape juice. Food Funct. 2013;4(1):52-62. Epub 2012/09/11. doi: 10.1039/c2fo30151b. PubMed PMID: 22961385.
87. Jimenez-Giron A, Queipo-Ortuno MI, Boto-Ordonez M, Munoz-Gonzalez I, Sanchez-Patan F, Monagas M, Martin-Alvarez PJ, Murri M, Tinahones FJ, Andres-Lacueva C, Bartolome B, Moreno-Arribas MV. Comparative study of microbial-derived phenolic metabolites in human feces after intake of gin, red wine, and dealcoholized red wine. J Agric Food Chem. 2013;61(16):3909-15. Epub 2013/04/13. doi: 10.1021/jf400678d. PubMed PMID: 23578197.
88. Gross G, Jacobs DM, Peters S, Possemiers S, van Duynhoven J, Vaughan EE, van de Wiele T. In vitro bioconversion of polyphenols from black tea and red wine/grape juice by human intestinal microbiota displays strong interindividual variability. J Agric Food Chem. 2010;58(18):10236-46. Epub 2010/08/24. doi: 10.1021/jf101475m. PubMed PMID: 20726519.
89. Bresciani L, Angelino D, Vivas EI, Kerby RL, Garcia-Viguera C, Del Rio D, Rey FE, Mena P. Differential Catabolism of an Anthocyanin-Rich Elderberry Extract by Three Gut Microbiota Bacterial Species. J Agric Food Chem. 2019. Epub 2019/04/11. doi: 10.1021/acs.jafc.9b00247. PubMed PMID: 30969770.
90. Carry E, Zhao D, Mogno I, Faith J, Ho L, Villani T, Patel H, Pasinetti GM, Simon JE, Wu Q. Targeted analysis of microbial-generated phenolic acid metabolites derived from grape flavanols by gas chromatography-triple quadrupole mass spectrometry. J Pharm Biomed Anal. 2018;159:374-83. Epub 2018/07/23. doi: 10.1016/j.jpba.2018.06.034. PubMed PMID: 30032004.
91. Ou K, Sarnoski P, Schneider KR, Song K, Khoo C, Gu L. Microbial catabolism of procyanidins by human gut microbiota. Mol Nutr Food Res. 2014;58(11):2196-205. Epub 2014/07/22. doi: 10.1002/mnfr.201400243. PubMed PMID: 25045165.
92. Jaganath IB, Mullen W, Lean ME, Edwards CA, Crozier A. In vitro catabolism of rutin by human fecal bacteria and the antioxidant capacity of its catabolites. Free Radic Biol Med. 2009;47(8):1180-9. Epub 2009/08/04. doi: 10.1016/j.freeradbiomed.2009.07.031. PubMed PMID: 19647790.
93. Bazzocco S, Mattila I, Guyot S, Renard CM, Aura AM. Factors affecting the conversion of apple polyphenols to phenolic acids and fruit matrix to short-chain fatty acids by human faecal microbiota in vitro. Eur J Nutr. 2008;47(8):442-52. Epub 2008/10/22. doi: 10.1007/s00394-008-0747-2. PubMed PMID: 18931964.
94. Goedert JJ, Sampson JN, Moore SC, Xiao Q, Xiong X, Hayes RB, Ahn J, Shi J, Sinha R. Fecal metabolomics: assay performance and association with colorectal cancer. Carcinogenesis. 2014;35(9):2089-96. Epub 2014/07/20. doi: 10.1093/carcin/bgu131. PubMed PMID: 25037050; PMCID: PMC4146421.
95. Temellini A, Mogavero S, Giulianotti PC, Pietrabissa A, Mosca F, Pacifici GM. Conjugation of benzoic acid with glycine in human liver and kidney: a study on the interindividual variability. Xenobiotica. 1993;23(12):1427-33. Epub 1993/12/01. doi: 10.3109/00498259309059451. PubMed PMID: 8135043.
96. Strahl NR, Barr WH. Intestinal drug absorption and metabolism. 3. Glycine conjugation and accumulation of benzoic acid in rat intestinal tissue. J Pharm Sci. 1971;60(2):278-81. Epub 1971/02/01. doi: 10.1002/jps.2600600227. PubMed PMID: 5572455.
97. Badenhorst CP, Erasmus E, van der Sluis R, Nortje C, van Dijk AA. A new perspective on the importance of glycine conjugation in the metabolism of aromatic acids. Drug Metab Rev. 2014;46(3):343-61. Epub 2014/04/24. doi: 10.3109/03602532.2014.908903. PubMed PMID: 24754494.
98. Drasar BS, Hill MJ. Metabolism of Nitrogen Compounds. Human Intestinal Flora. London: Academic Press; 1974. p. 72-102.
99. Smith EA, Macfarlane GT. Enumeration of human colonic bacteria producing phenolic and indolic compounds: effects of pH, carbohydrate availability and retention time on dissimilatory aromatic amino acid metabolism. J Appl Bacteriol. 1996;81(3):288-302. Epub 1996/09/01. PubMed PMID: 8810056.
100. Smith EA, Macfarlane GT. Formation of Phenolic and Indolic Compounds by Anaerobic Bacteria in the Human Large Intestine. Microb Ecol. 1997;33(3):180-8. Epub 1997/04/01. PubMed PMID: 9115181.
101. Bone E, Tamm A, Hill M. The production of urinary phenols by gut bacteria and their possible role in the causation of large bowel cancer. Am J Clin Nutr. 1976;29(12):1448-54.
102. Saito Y, Sato T, Nomoto K, Tsuji H. Identification of phenol- and p-cresol-producing intestinal bacteria by using media supplemented with tyrosine and its metabolites. FEMS Microbiol Ecol. 2018;94(9). Epub 2018/07/10. doi: 10.1093/femsec/fiy125. PubMed PMID: 29982420; PMCID: PMC6424909.
103. Jacobs DM, Deltimple N, van Velzen E, van Dorsten FA, Bingham M, Vaughan EE, van Duynhoven J. 1H NMR metabolite profiling of feces as a tool to assess the impact of nutrition on the human microbiome. NMR Biomed. 2008;21(6):615-26. doi: 10.1002/nbm.1233.
104. Smith EA, Macfarlane GT. Dissimilatory amino acid metabolism in human colonic bacteria. Anaerobe. 1997;3(5):327-37. Epub 2006/08/05. doi: 10.1006/anae.1997.0121. PubMed PMID: 16887608.
105. de Loor H, Bammens B, Evenepoel P, De Preter V, Verbeke K. Gas chromatographic-mass spectrometric analysis for measurement of p-cresol and its conjugated metabolites in uremic and normal serum. Clin Chem. 2005;51(8):1535-8. Epub 2005/07/26. doi: 10.1373/clinchem.2005.050781. PubMed PMID: 16040852.
106. Kobashi K, Fukaya Y, Kim DH, Akao T, Takebe S. A novel type of aryl sulfotransferase obtained from an anaerobic bacterium of human intestine. Arch Biochem Biophys. 1986;245(2):537-9. Epub 1986/03/01. doi: 10.1016/0003-9861(86)90247-x. PubMed PMID: 3456739.
107. Kim DH, Kobashi K. The role of intestinal flora in metabolism of phenolic sulfate esters. Biochem Pharmacol. 1986;35(20):3507-10. Epub 1986/10/15. doi: 10.1016/0006-2952(86)90619-2. PubMed PMID: 3464281.
108. Sundaram RS, Szumlanski C, Otterness D, van Loon JA, Weinshilboum RM. Human intestinal phenol sulfotransferase: assay conditions, activity levels and partial purification of the thermolabile form. Drug Metab Dispos. 1989;17(3):255-64. Epub 1989/05/01. PubMed PMID: 2568905.
109. Riches Z, Stanley EL, Bloomer JC, Coughtrie MW. Quantitative evaluation of the expression and activity of five major sulfotransferases (SULTs) in human tissues: the SULT “pie”. Drug Metab Dispos. 2009;37(11):2255-61. Epub 2009/08/15. doi: 10.1124/dmd.109.028399. PubMed PMID: 19679676; PMCID: PMC2774979.
110. Coughtrie MWH. Function and organization of the human cytosolic sulfotransferase (SULT) family. Chem Biol Interact. 2016;259(Pt A):2-7. Epub 2016/05/14. doi: 10.1016/j.cbi.2016.05.005. PubMed PMID: 27174136.
111. Vijay N, Morris ME. Role of monocarboxylate transporters in drug delivery to the brain. Curr Pharm Des. 2014;20(10):1487-98. Epub 2013/06/25. doi: 10.2174/13816128113199990462. PubMed PMID: 23789956; PMCID: PMC4084603.
112. Kobayashi S, Konishi Y. Transepithelial transport of flavanone in intestinal Caco-2 cell monolayers. Biochem Biophys Res Commun. 2008;368(1):23-9. Epub 2008/01/15. doi: 10.1016/j.bbrc.2007.12.185. PubMed PMID: 18190788.
113. Hoverstad T, Carlstedt-Duke B, Lingaas E, Norin E, Saxerholt H, Steinbakk M, Midtvedt T. Influence of oral intake of seven different antibiotics on faecal short-chain fatty acid excretion in healthy subjects. Scand J Gastroenterol. 1986;21(8):997-1003. Epub 1986/10/01. doi: 10.3109/00365528608996411. PubMed PMID: 3775265.
114. Borthakur A, Priyamvada S, Kumar A, Natarajan AA, Gill RK, Alrefai WA, Dudeja PK. A novel nutrient sensing mechanism underlies substrate-induced regulation of monocarboxylate transporter-1. Am J Physiol Gastrointest Liver Physiol. 2012;303(10):G1126-33. Epub 2012/09/18. doi: 10.1152/ajpgi.00308.2012. PubMed PMID: 22982338; PMCID: PMC3517653.
115. Gryp T, De Paepe K, Vanholder R, Kerckhof FM, Van Biesen W, Van de Wiele T, Verbeke F, Speeckaert M, Joossens M, Couttenye MM, Vaneechoutte M, Glorieux G. Gut microbiota generation of protein-bound uremic toxins and related metabolites is not altered at different stages of chronic kidney disease. Kidney Int. 2020. Epub 2020/04/23. doi: 10.1016/j.kint.2020.01.028. PubMed PMID: 32317112.
116. Loftfield E, Vogtmann E, Sampson JN, Moore SC, Nelson H, Knight R, Chia N, Sinha R. Comparison of Collection Methods for Fecal Samples for Discovery Metabolomics in Epidemiologic Studies. Cancer Epidemiol Biomarkers Prev. 2016;25(11):1483-90. Epub 2016/11/03. doi: 10.1158/1055-9965.EPI-16-0409. PubMed PMID: 27543620; PMCID: PMC5093035.
117. Mutsaers HA, Caetano-Pinto P, Seegers AE, Dankers AC, van den Broek PH, Wetzels JF, van den Brand JA, van den Heuvel LP, Hoenderop JG, Wilmer MJ, Masereeuw R. Proximal tubular efflux transporters involved in renal excretion of p-cresyl sulfate and p-cresyl glucuronide: Implications for chronic kidney disease pathophysiology. Toxicol In Vitro. 2015;29(7):1868-77. Epub 2015/07/29. doi: 10.1016/j.tiv.2015.07.020. PubMed PMID: 26216510.
118. Maliepaard M, Scheffer GL, Faneyte IF, van Gastelen MA, Pijnenborg AC, Schinkel AH, van De Vijver MJ, Scheper RJ, Schellens JH. Subcellular localization and distribution of the breast cancer resistance protein transporter in normal human tissues. Cancer Res. 2001;61(8):3458-64. Epub 2001/04/20. PubMed PMID: 11309308.
119. Seithel A, Karlsson J, Hilgendorf C, Bjorquist A, Ungell AL. Variability in mRNA expression of ABC- and SLC-transporters in human intestinal cells: comparison between human segments and Caco-2 cells. Eur J Pharm Sci. 2006;28(4):291-9. Epub 2006/05/18. doi: 10.1016/j.ejps.2006.03.003. PubMed PMID: 16704924.
120. Miyamoto Y, Watanabe H, Noguchi T, Kotani S, Nakajima M, Kadowaki D, Otagiri M, Maruyama T. Organic anion transporters play an important role in the uptake of p-cresyl sulfate, a uremic toxin, in the kidney. Nephrol Dial Transplant. 2011;26(8):2498-502. Epub 2011/02/10. doi: 10.1093/ndt/gfq785. PubMed PMID: 21303967.
121. Mutsaers HA, Wilmer MJ, Reijnders D, Jansen J, van den Broek PH, Forkink M, Schepers E, Glorieux G, Vanholder R, van den Heuvel LP, Hoenderop JG, Masereeuw R. Uremic toxins inhibit renal metabolic capacity through interference with glucuronidation and mitochondrial respiration. Biochim Biophys Acta. 2013;1832(1):142-50. Epub 2012/09/29. doi: 10.1016/j.bbadis.2012.09.006. PubMed PMID: 23017367.
122. Watanabe H, Sakaguchi Y, Sugimoto R, Kaneko K, Iwata H, Kotani S, Nakajima M, Ishima Y, Otagiri M, Maruyama T. Human organic anion transporters function as a high-capacity transporter for p-cresyl sulfate, a uremic toxin. Clin Exp Nephrol. 2014;18(5):814-20. Epub 2013/11/05. doi: 10.1007/s10157-013-0902-9. PubMed PMID: 24185403.
123. Peng YH, Sweet DH, Lin SP, Yu CP, Lee Chao PD, Hou YC. Green tea inhibited the elimination of nephro-cardiovascular toxins and deteriorated the renal function in rats with renal failure. Sci Rep. 2015;5:16226. Epub 2015/11/11. doi: 10.1038/srep16226. PubMed PMID: 26552961; PMCID: PMC4639770.
124. Hsueh CH, Yoshida K, Zhao P, Meyer TW, Zhang L, Huang SM, Giacomini KM. Identification and Quantitative Assessment of Uremic Solutes as Inhibitors of Renal Organic Anion Transporters, OAT1 and OAT3. Mol Pharm. 2016;13(9):3130-40. Epub 2016/07/29. doi: 10.1021/acs.molpharmaceut.6b00332. PubMed PMID: 27467266.
125. Sun W, Wu RR, van Poelje PD, Erion MD. Isolation of a family of organic anion transporters from human liver and kidney. Biochem Biophys Res Commun. 2001;283(2):417-22. Epub 2001/05/01. doi: 10.1006/bbrc.2001.4774. PubMed PMID: 11327718.
126. Hilgendorf C, Ahlin G, Seithel A, Artursson P, Ungell AL, Karlsson J. Expression of thirty-six drug transporter genes in human intestine, liver, kidney, and organotypic cell lines. Drug Metab Dispos. 2007;35(8):1333-40. Epub 2007/05/15. doi: 10.1124/dmd.107.014902. PubMed PMID: 17496207.
127. Nicol MR, Fedoriw Y, Mathews M, Prince HM, Patterson KB, Geller E, Mollan K, Mathews S, Kroetz DL, Kashuba AD. Expression of six drug transporters in vaginal, cervical, and colorectal tissues: Implications for drug disposition in HIV prevention. J Clin Pharmacol. 2014;54(5):574-83. Epub 2013/12/18. doi: 10.1002/jcph.248. PubMed PMID: 24343710; PMCID: PMC4061289.
128. Jariyawat S, Sekine T, Takeda M, Apiwattanakul N, Kanai Y, Sophasan S, Endou H. The interaction and transport of beta-lactam antibiotics with the cloned rat renal organic anion transporter 1. J Pharmacol Exp Ther. 1999;290(2):672-7. Epub 1999/07/20. PubMed PMID: 10411577.
129. Takeda M, Babu E, Narikawa S, Endou H. Interaction of human organic anion transporters with various cephalosporin antibiotics. Eur J Pharmacol. 2002;438(3):137-42. Epub 2002/03/23. doi: 10.1016/s0014-2999(02)01306-7. PubMed PMID: 11909604.
130. Akiki TJ, Averill CL, Wrocklage KM, Schweinsburg B, Scott JC, Martini B, Averill LA, Southwick SM, Krystal JH, Abdallah CG. The Association of PTSD Symptom Severity with Localized Hippocampus and Amygdala Abnormalities. Chronic Stress (Thousand Oaks). 2017;1. Epub 2017/08/22. doi: 10.1177/2470547017724069. PubMed PMID: 28825050; PMCID: PMC5562232.
131. Luo SS, Yu CP, Hsieh YW, Chao PL, Sweet DH, Hou YC, Lin SP. Effects of antibiotics on the pharmacokinetics of indoxyl sulfate, a nephro-cardiovascular toxin. Xenobiotica. 2020;50(5):588-92. Epub 2019/08/27. doi: 10.1080/00498254.2019.1660433. PubMed PMID: 31448977.
132. DeMoss RD, Moser K. Tryptophanase in diverse bacterial species. J Bacteriol. 1969;98(1):167-71. Epub 1969/04/01. PubMed PMID: 5781572; PMCID: PMC249919.
133. Kaur H, Bose C, Mande SS. Tryptophan Metabolism by Gut Microbiome and Gut-Brain-Axis: An in silico Analysis. Front Neurosci. 2019;13:1365. Epub 2020/01/11. doi: 10.3389/fnins.2019.01365. PubMed PMID: 31920519; PMCID: PMC6930238.
134. Amaretti A, Gozzoli C, Simone M, Raimondi S, Righini L, Perez-Brocal V, Garcia-Lopez R, Moya A, Rossi M. Profiling of Protein Degraders in Cultures of Human Gut Microbiota. Front Microbiol. 2019;10:2614. Epub 2019/12/06. doi: 10.3389/fmicb.2019.02614. PubMed PMID: 31803157; PMCID: PMC6874058.
135. Darkoh C, Chappell C, Gonzales C, Okhuysen P. A rapid and specific method for the detection of indole in complex biological samples. Appl Environ Microbiol. 2015;81(23):8093-7. Epub 2015/09/20. doi: 10.1128/aem.02787-15. PubMed PMID: 26386049; PMCID: PMC4651089.
136. Banoglu E, Jha GG, King RS. Hepatic microsomal metabolism of indole to indoxyl, a precursor of indoxyl sulfate. Eur J Drug Metab Pharmacokinet. 2001;26(4):235-40. Epub 2002/01/26. doi: 10.1007/bf03226377. PubMed PMID: 11808865; PMCID: PMC2254176.
137. Gillam EM, Notley LM, Cai H, De Voss JJ, Guengerich FP. Oxidation of indole by cytochrome P450 enzymes. Biochemistry. 2000;39(45):13817-24. Epub 2000/11/15. doi: 10.1021/bi001229u. PubMed PMID: 11076521.
138. Banoglu E, King RS. Sulfation of indoxyl by human and rat aryl (phenol) sulfotransferases to form indoxyl sulfate. Eur J Drug Metab Pharmacokinet. 2002;27(2):135-40. Epub 2002/06/18. doi: 10.1007/bf03190428. PubMed PMID: 12064372; PMCID: PMC2254172.
139. King LJ, Parke DV, Williams RT. The metabolism of [2-14C] indole in the rat. Biochem J. 1966;98(1):266-77. Epub 1966/01/01. doi: 10.1042/bj0980266. PubMed PMID: 5328168; PMCID: PMC1264825.
140. Morimoto K, Tominaga Y, Agatsuma Y, Miyamoto M, Kashiwagura S, Takahashi A, Sano Y, Yano K, Kakinuma C, Ogihara T, Tomita M. Intestinal secretion of indoxyl sulfate as a possible compensatory excretion pathway in chronic kidney disease. Biopharm Drug Dispos. 2018;39(7):328-34. Epub 2018/07/06. doi: 10.1002/bdd.2149. PubMed PMID: 29975986.
141. Konopelski P, Konop M, Gawrys-Kopczynska M, Podsadni P, Szczepanska A, Ufnal M. Indole-3-Propionic Acid, a Tryptophan-Derived Bacterial Metabolite, Reduces Weight Gain in Rats. Nutrients. 2019;11(3). Epub 2019/03/14. doi: 10.3390/nu11030591. PubMed PMID: 30862081; PMCID: PMC6471155.
142. Gertsman I, Gangoiti JA, Nyhan WL, Barshop BA. Perturbations of tyrosine metabolism promote the indolepyruvate pathway via tryptophan in host and microbiome. Mol Genet Metab. 2015;114(3):431-7. Epub 2015/02/15. doi: 10.1016/j.ymgme.2015.01.005. PubMed PMID: 25680927.
143. Dodd D, Spitzer MH, Van Treuren W, Merrill BD, Hryckowian AJ, Higginbottom SK, Le A, Cowan TM, Nolan GP, Fischbach MA, Sonnenburg JL. A gut bacterial pathway metabolizes aromatic amino acids into nine circulating metabolites. Nature. 2017;551(7682):648-52. Epub 2017/11/24. doi: 10.1038/nature24661. PubMed PMID: 29168502; PMCID: PMC5850949.
144. Elsden SR, Hilton MG, Waller JM. The end products of the metabolism of aromatic amino acids by Clostridia. Arch Microbiol. 1976;107(3):283-8. Epub 1976/04/01. PubMed PMID: 1275638.
145. Elsden SR, Hilton MG. Amino acid utilization patterns in clostridial taxonomy. Arch Microbiol. 1979;123(2):137-41. Epub 1979/11/01. PubMed PMID: 539867.
146. Jellet JJ, Forrest TP, Macdonald IA, Marrie TJ, Holdeman LV. Production of indole-3-propanoic acid and 3-(p-hydroxyphenyl)propanoic acid by Clostridium sporogenes: a convenient thin-layer chromatography detection system. Can J Microbiol. 1980;26(4):448-53. Epub 1980/04/01. doi: 10.1139/m80-074. PubMed PMID: 7378938.
147. Wlodarska M, Luo C, Kolde R, d’Hennezel E, Annand JW, Heim CE, Krastel P, Schmitt EK, Omar AS, Creasey EA, Garner AL, Mohammadi S, O’Connell DJ, Abubucker S, Arthur TD, Franzosa EA, Huttenhower C, Murphy LO, Haiser HJ, Vlamakis H, Porter JA, Xavier RJ. Indoleacrylic Acid Produced by Commensal Peptostreptococcus Species Suppresses Inflammation. Cell Host Microbe. 2017;22(1):25-37.e6. Epub 2017/07/14. doi: 10.1016/j.chom.2017.06.007. PubMed PMID: 28704649; PMCID: PMC5672633.
148. Pavlova T, Vidova V, Bienertova-Vasku J, Janku P, Almasi M, Klanova J, Spacil Z. Urinary intermediates of tryptophan as indicators of the gut microbial metabolism. Anal Chim Acta. 2017;987:72-80. Epub 2017/09/17. doi: 10.1016/j.aca.2017.08.022. PubMed PMID: 28916042.
149. Metzner L, Kottra G, Neubert K, Daniel H, Brandsch M. Serotonin, L-tryptophan, and tryptamine are effective inhibitors of the amino acid transport system PAT1. FASEB J. 2005;19(11):1468-73. Epub 2005/08/30. doi: 10.1096/fj.05-3683com. PubMed PMID: 16126914.
150. Roshchina VV. Evolutionary Considerations of Neurotransmitters in Microbial, Plant, and Animal Cells. In: Lyte M, Freestone PPE, editors. Microbial Endocrinology: Interkingdom Signaling in Infectious Disease and Health. New York, NY: Springer New York; 2010. p. 17-52.
151. Erspamer V. Occurrence of indolealkylamines in nature. In: Erspamer V, editor. 5-Hydroxytryptamine and Related Indolealkylamines. Berlin, Heidelberg: Springer Berlin Heidelberg; 1966. p. 132-81.
152. Bertaccini G. Tissue 5-hydroxytryptamine and urinary 5-hydroxyindoleacetic acid after partial or total removal of the gastro-intestinal tract in the rat. J Physiol. 1960;153(2):239-49. Epub 1960/09/01. doi: 10.1113/jphysiol.1960.sp006532. PubMed PMID: 16992061; PMCID: PMC1359746.
153. Yano JM, Yu K, Donaldson GP, Shastri GG, Ann P, Ma L, Nagler CR, Ismagilov RF, Mazmanian SK, Hsiao EY. Indigenous bacteria from the gut microbiota regulate host serotonin biosynthesis. Cell. 2015;161(2):264-76. Epub 2015/04/11. doi: 10.1016/j.cell.2015.02.047. PubMed PMID: 25860609; PMCID: PMC4393509.
154. Reigstad CS, Salmonson CE, Rainey JF, 3rd, Szurszewski JH, Linden DR, Sonnenburg JL, Farrugia G, Kashyap PC. Gut microbes promote colonic serotonin production through an effect of short-chain fatty acids on enterochromaffin cells. FASEB J. 2015;29(4):1395-403. Epub 2015/01/01. doi: 10.1096/fj.14-259598. PubMed PMID: 25550456; PMCID: PMC4396604.
155. Cummings JH. Short chain fatty acids. In: Gibson GR, Macfarlane GT, editors. Human Colonic Bacteria : Role in Nutrition, Physiology and Pathology. Boca Raton, FL: CRC Press; 1995. p. 101-30.
156. Hoverstad T, Midtvedt T. Short-chain fatty acids in germfree mice and rats. J Nutr. 1986;116(9):1772-6. Epub 1986/09/01. doi: 10.1093/jn/116.9.1772. PubMed PMID: 3761032.
157. Perry RJ, Peng L, Barry NA, Cline GW, Zhang D, Cardone RL, Petersen KF, Kibbey RG, Goodman AL, Shulman GI. Acetate mediates a microbiome-brain-beta-cell axis to promote metabolic syndrome. Nature. 2016;534(7606):213-7. Epub 2016/06/10. doi: 10.1038/nature18309. PubMed PMID: 27279214; PMCID: PMC4922538.
158. Deshpande A, Hurless K, Cadnum JL, Chesnel L, Gao L, Chan L, Kundrapu S, Polinkovsky A, Donskey CJ. Effect of Fidaxomicin versus Vancomycin on Susceptibility to Intestinal Colonization with Vancomycin-Resistant Enterococci and Klebsiella pneumoniae in Mice. Antimicrob Agents Chemother. 2016;60(7):3988-93. Epub 2016/04/20. doi: 10.1128/aac.02590-15. PubMed PMID: 27090175; PMCID: PMC4914684.
159. Ahmed S, Macfarlane GT, Fite A, McBain AJ, Gilbert P, Macfarlane S. Mucosa-associated bacterial diversity in relation to human terminal ileum and colonic biopsy samples. Appl Environ Microbiol. 2007;73(22):7435-42. Epub 2007/09/25. doi: 10.1128/aem.01143-07. PubMed PMID: 17890331; PMCID: PMC2168195.
160. Simon GL, Gorbach SL. Intestinal flora in health and disease. Gastroenterology. 1984;86(1):174-93. doi: 10.1016/0016-5085(84)90606-1.
161. Laux DC, Cohen PS, Conway T. Role of the Mucus Layer in Bacterial Colonization of the Intestine. In: Nataro J, Cohen P, Mobley H, Weiser J, editors. Colonization of Mucosal Surfaces. Washington, DC: ASM Press; 2005. p. 119-212.
162. Zoetendal EG, Raes J, van den Bogert B, Arumugam M, Booijink CC, Troost FJ, Bork P, Wels M, de Vos WM, Kleerebezem M. The human small intestinal microbiota is driven by rapid uptake and conversion of simple carbohydrates. The ISME journal. 2012;6(7):1415-26. Epub 2012/01/20. doi: 10.1038/ismej.2011.212. PubMed PMID: 22258098; PMCID: PMC3379644.
163. Schantz M, Erk T, Richling E. Metabolism of green tea catechins by the human small intestine. Biotechnol J. 2010;5(10):1050-9. Epub 2010/10/12. doi: 10.1002/biot.201000214. PubMed PMID: 20931601.
164. van Kessel SP, Frye AK, El-Gendy AO, Castejon M, Keshavarzian A, van Dijk G, El Aidy S. Gut bacterial tyrosine decarboxylases restrict levels of levodopa in the treatment of Parkinson’s disease. Nature communications. 2019;10(1):310. Epub 2019/01/20. doi: 10.1038/s41467-019-08294-y. PubMed PMID: 30659181; PMCID: PMC6338741.
165. Aronov PA, Luo FJ, Plummer NS, Quan Z, Holmes S, Hostetter TH, Meyer TW. Colonic contribution to uremic solutes. J Am Soc Nephrol. 2011;22(9):1769-76. Epub 2011/07/26. doi: 10.1681/ASN.2010121220. PubMed PMID: 21784895; PMCID: PMC3171947.
166. Mair RD, Sirich TL, Plummer NS, Meyer TW. Characteristics of Colon-Derived Uremic Solutes. Clin J Am Soc Nephrol. 2018;13(9):1398-404. Epub 2018/08/09. doi: 10.2215/cjn.03150318. PubMed PMID: 30087103; PMCID: PMC6140561.
167. Willmann M, Vehreschild M, Biehl LM, Vogel W, Dorfel D, Hamprecht A, Seifert H, Autenrieth IB, Peter S. Distinct impact of antibiotics on the gut microbiome and resistome: a longitudinal multicenter cohort study. BMC Biol. 2019;17(1):76. Epub 2019/09/20. doi: 10.1186/s12915-019-0692-y. PubMed PMID: 31533707; PMCID: PMC6749691.
168. Farowski F, Els G, Tsakmaklis A, Higgins PG, Kahlert CR, Stein-Thoeringer CK, Bobardt JS, Dettmer-Wilde K, Oefner PJ, Vehreschild JJ, Vehreschild M. Assessment of urinary 3-indoxyl sulfate as a marker for gut microbiota diversity and abundance of Clostridiales. Gut microbes. 2019;10(2):133-41. Epub 2018/08/18. doi: 10.1080/19490976.2018.1502536. PubMed PMID: 30118620; PMCID: PMC6546351.
169. Rashid MU, Zaura E, Buijs MJ, Keijser BJ, Crielaard W, Nord CE, Weintraub A. Determining the Long-term Effect of Antibiotic Administration on the Human Normal Intestinal Microbiota Using Culture and Pyrosequencing Methods. Clin Infect Dis. 2015;60 Suppl 2:S77-84. Epub 2015/04/30. doi: 10.1093/cid/civ137. PubMed PMID: 25922405.
170. Segal LN, Clemente JC, Wu BG, Wikoff WR, Gao Z, Li Y, Ko JP, Rom WN, Blaser MJ, Weiden MD. Randomised, double-blind, placebo-controlled trial with azithromycin selects for anti-inflammatory microbial metabolites in the emphysematous lung. Thorax. 2017;72(1):13-22. Epub 2016/08/04. doi: 10.1136/thoraxjnl-2016-208599. PubMed PMID: 27486204; PMCID: PMC5329050.
171. Pérez-Cobas AE, Artacho A, Knecht H, Ferrús ML, Friedrichs A, Ott SJ, Moya A, Latorre A, Gosalbes MJ. Differential effects of antibiotic therapy on the structure and function of human gut microbiota. PLoS One. 2013;8(11):e80201. Epub 2013/11/28. doi: 10.1371/journal.pone.0080201. PubMed PMID: 24282523; PMCID: PMC3839934.
172. Dethlefsen L, Huse S, Sogin ML, Relman DA. The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol. 2008;6(11):e280. Epub 2008/11/21. doi: 10.1371/journal.pbio.0060280. PubMed PMID: 19018661; PMCID: PMC2586385.
173. Macfarlane S, Macfarlane GT. Proteolysis and amino acid fermentation. Human Colonic Bacteria: Role in Nutrition, Physiology, and Pathology. Boca Raton, FL: CRC Press; 1995. p. 75-100.
174. Nunez-Montiel OL, Thompson FS, Dowell VR, Jr. Norleucine-tyrosine broth for rapid identification of Clostridium difficile by gas-liquid chromatography. J Clin Microbiol. 1983;17(2):382-5. Epub 1983/02/01. PubMed PMID: 6833488; PMCID: PMC272645.
175. Darkoh C, Plants-Paris K, Bishoff D, DuPont HL. Clostridium difficile Modulates the Gut Microbiota by Inducing the Production of Indole, an Interkingdom Signaling and Antimicrobial Molecule. mSystems. 2019;4(2). Epub 2019/04/05. doi: 10.1128/mSystems.00346-18. PubMed PMID: 30944877; PMCID: PMC6426650.
176. Winter J, Popoff MR, Grimont P, Bokkenheuser VD. Clostridium orbiscindens sp. nov., a human intestinal bacterium capable of cleaving the flavonoid C-ring. Int J Syst Bacteriol. 1991;41(3):355-7. Epub 1991/07/01. doi: 10.1099/00207713-41-3-355. PubMed PMID: 1883711.
177. Schoefer L, Mohan R, Schwiertz A, Braune A, Blaut M. Anaerobic degradation of flavonoids by Clostridium orbiscindens. Appl Environ Microbiol. 2003;69(10):5849-54. Epub 2003/10/09. doi: 10.1128/aem.69.10.5849-5854.2003. PubMed PMID: 14532034; PMCID: PMC201214.
178. Peng X, Zhang Z, Zhang N, Liu L, Li S, Wei H. In vitro catabolism of quercetin by human fecal bacteria and the antioxidant capacity of its catabolites. Food Nutr Res. 2014;58. Epub 2014/04/26. doi: 10.3402/fnr.v58.23406. PubMed PMID: 24765061; PMCID: PMC3991839.
179. Hur HG, Beger RD, Heinze TM, Lay JO, Jr., Freeman JP, Dore J, Rafii F. Isolation of an anaerobic intestinal bacterium capable of cleaving the C-ring of the isoflavonoid daidzein. Arch Microbiol. 2002;178(1):8-12. Epub 2002/06/19. doi: 10.1007/s00203-002-0414-6. PubMed PMID: 12070764.
180. van Duynhoven J, van der Hooft JJ, van Dorsten FA, Peters S, Foltz M, Gomez-Roldan V, Vervoort J, de Vos RC, Jacobs DM. Rapid and sustained systemic circulation of conjugated gut microbial catabolites after single-dose black tea extract consumption. J Proteome Res. 2014;13(5):2668-78. Epub 2014/03/29. doi: 10.1021/pr5001253. PubMed PMID: 24673575.
181. Law WS, Huang PY, Ong ES, Ong CN, Li SF, Pasikanti KK, Chan EC. Metabonomics investigation of human urine after ingestion of green tea with gas chromatography/mass spectrometry, liquid chromatography/mass spectrometry and (1)H NMR spectroscopy. Rapid Commun Mass Spectrom. 2008;22(16):2436-46. Epub 2008/07/18. doi: 10.1002/rcm.3629. PubMed PMID: 18634125.
182. Jacobs DM, Fuhrmann JC, van Dorsten FA, Rein D, Peters S, van Velzen EJ, Hollebrands B, Draijer R, van Duynhoven J, Garczarek U. Impact of short-term intake of red wine and grape polyphenol extract on the human metabolome. J Agric Food Chem. 2012;60(12):3078-85. Epub 2012/03/01. doi: 10.1021/jf2044247. PubMed PMID: 22372405.
183. Ling WH, Hänninen O. Shifting from a conventional diet to an uncooked vegan diet reversibly alters fecal hydrolytic activities in humans. J Nutr. 1992;122(4):924-30. Epub 1992/04/01. doi: 10.1093/jn/122.4.924. PubMed PMID: 1552366.
184. Barrios C, Beaumont M, Pallister T, Villar J, Goodrich JK, Clark A, Pascual J, Ley RE, Spector TD, Bell JT, Menni C. Gut-Microbiota-Metabolite Axis in Early Renal Function Decline. PLoS One. 2015;10(8):e0134311. Epub 2015/08/05. doi: 10.1371/journal.pone.0134311. PubMed PMID: 26241311; PMCID: PMC4524635.
185. Ichinohe T, Pang IK, Kumamoto Y, Peaper DR, Ho JH, Murray TS, Iwasaki A. Microbiota regulates immune defense against respiratory tract influenza A virus infection. Proc Natl Acad Sci U S A. 2011;108(13):5354-9. Epub 2011/03/16. doi: 10.1073/pnas.1019378108. PubMed PMID: 21402903; PMCID: PMC3069176.
186. Jaskiw GE, Obrenovich ME, Donskey CJ. The phenolic interactome and gut microbiota: opportunities and challenges in developing applications for schizophrenia and autism. Psychopharmacology (Berl). 2019;236(5):1471-89. Epub 2019/06/15. doi: http://dx.doi.org/10.1007/s00213-019-05267-3. PubMed PMID: 31197432.
Submitted June 16, 2020 | Accepted October 16, 2020 | Published December 29, 2020
Copyright © 2020 Pathogens and Immunity. This is an open-access article distributed under the terms of the Creative Commons Attribution 4.0 International License.