Carol L. Vinton1, Carly E. Starke1, Alexandra M. Ortiz1, Stephen H. Lai1, Jacob K. Flynn1, Ornella Sortino2, Kenneth Knox3, Irini Sereti2, Jason M. Brenchley1
1Barrier Immunity Section; Laboratory of Viral Diseases; NIAID, NIH; Bethesda, Maryland
2HIV Pathogenesis Section; Laboratory of Immunoregulation; NIAID, NIH; Bethesda, Maryland
3Department of Medicine; University of Arizona; Tucson, Arizona
Jason M. Brenchley
National Institute of Allergy and Infectious Diseases
4 Memorial Drive, Bldg. 4/Rm. 211
Bethesda, MD 20892
Phone: 301-496-1498
jbrenchl@niaid.nih.gov
Background: Microbial translocation occurs after damage to the structural and/or immunological barrier of the gastrointestinal (GI) tract into circulation. Microbial components that translocate from the lumen of the GI tract directly stimulate the immune system and contribute to inflammation. When microbial translocation becomes chronic, the inflammation has detrimental consequences. Given that microbial translocation is an important phenomenon in many diseases, defining biomarkers that reliably reflect microbial translocation is critical. Measurement of systemic microbial products is difficult since: 1) robust assays to measure microbial antigens simultaneously are lacking; 2) confounding factors influence assays used to detect microbial products; and 3) biological clearance mechanisms limit their detection in circulation. Thus, host proteins produced in response to microbial stimulation are used as surrogates for microbial translocation; however, many of these proteins are also produced in response to host proteins expressed by dying cells.
Methods: We measured plasma levels of biomarkers associated with GI tract damage, immune responses to microbial products, and cell-death in people living with HIV before and after antiretroviral administration, and in macaque nonhuman primates before and after SIV infection.
Results: Proteins secreted during cellular stress (receptor for advanced glycation endproducts - RAGE and high motility group box 1 -HMGB1), which can induce sCD14 production in vitro and in vivo, do not associate with elevated levels of biomarkers associated with microbial translocation in progressively HIV-infected individuals and SIV-infected NHPs.
Conclusions: Bystander cell death and generalized inflammation do not contribute to elevated levels of sCD14 observed in HIV/SIV-infected individuals.
Keywords: Microbial translocation; HIV; SIV; inflammation
Damage to the immunological and structural barrier of the gastrointestinal (GI) tract which occurs during acute HIV infection of humans and SIV infection of Asian macaques leads to translocation of microbial products from the lumen of the GI tract into circulation [1-3]. Significant loss of GI tract-resident CD4+ T cells [4], decreased production of IL-17 and IL-22 effector cytokines by GI tract-resident lymphocytes [5-7], alteration to the landscape of antigen presenting cells [5, 8], and epithelial cell apoptosis with loss of epithelial integrity [2,9, 10] all contribute to epithelial barrier damage and, thus, microbial translocation in HIV/SIV infected individuals. Microbial products directly stimulate the immune system and, therefore, contribute to systemic inflammation in immunodeficiency lentiviral infections. While microbial translocation is one cause of systemic inflammation, the degree to which microbial translocation contributes to inflammation relative to HIV/SIV replication, and to frequently co-existing co-infections, in immunocompromised individuals is not fully known. The contribution of HIV/SIV to inflammation can be assessed, to a large degree, by treating people living with HIV (PLWH) and SIV-infected NHPs with antiretroviral therapy (ART). Indeed, after administration of ART systemic inflammation decreases but remains elevated relative to healthy humans and SIV-uninfected NHPs. Residual inflammation observed in ART-treated PLWH and SIV-infected NHPs is associated with increased morbidity and mortality as compared to uninfected, population controls [11-14]. Thus, there is great interest in understanding the contribution of microbial translocation to this residual inflammation.
Measurement of the degree of microbial translocation occurring in vivo has been difficult. Bacterial DNA can be detected by quantitative PCR using primers which detect conserved sequences in the DNA that encode bacterial ribosomal RNA and elevated levels of bacterial DNA have been observed in PLWH [15-17]. The sensitivity of this assay is very low; however extracellular endonucleases in plasma and tissues can degrade extracellular DNA [18]. Additionally, bacterial DNA contamination is common in biomedical science which can confound the assay. The bacterial cell wall component lipopolysaccharide (LPS) can also be measured in biological materials by the limulus amebocyte lysate (LAL) assay. This assay takes advantage of an enzymatic reaction within the horseshoe crab (limulus) coagulation system that occurs in the presence of LPS. LPS levels are detectable in the pg/ml range with the LAL assay [1]. However, the ability of this assay to reliably detect LPS is greatly influenced by the presence of plasma proteins, such as high-density lipoproteins that result in its hepatic clearance and natural antibodies that recognize LPS, both of which can shield LPS from the LAL enzymatic reactions [19]. Thus, the ability to reliably measure systemic bacterial DNA and plasma LPS is highly variable.
A third approach to ascertaining levels of microbial translocation involves immunohistochemical staining for microbial products in paraffin-embedded tissue sections [2]. This requires tissues obtained at necropsy or after invasive biopsy procedures but allows for unambiguous determination of where microbial products reside, whether they co-locate with proinflammatory mediators, how effectively they are being cleared by phagocytes, and can be used to precisely enumerate products of microbial translocation with quantitative image analysis [2]. However, given the amount of tissue required, this approach is not easily adapted to large cohorts, or to longitudinal studies. Thus, while there is considerable interest in understanding the causes and consequences of microbial translocation, assays for precise quantitative determination remain undefined.
As a surrogate to measuring microbial products directly, many studies measure host proteins made, predominantly, in response to microbial antigenic stimulation. Commonly, plasma levels of soluble CD14 (sCD14, produced by myeloid cells after stimulation by LPS), lipopolysaccharide-binding protein (LBP, produced by hepatocytes after LPS-stimulation), and/or natural antibodies directed against LPS core antigen (EndoCAb) [20] are used as surrogate biomarkers. Of these, sCD14 is most commonly measured as an estimate of the level of microbial translocation given that it can be easily measured in plasma [20]. With TLR4, CD14 is part of the LPS receptor. Myeloid cells that are stimulated with LPS produce and secrete sCD14 and circulating sCD14 levels correspond to the degree to which myeloid cells were stimulated with LPS in vivo [21]. However, LPS is not the only ligand for CD14/TLR4 and the degree to which sCD14 levels accurately reflect microbial translocation in PLWH has been disputed [22-24]. RAGE shares common ligands and signaling pathways with the CD14/TLR4 complex - including high mobility group box protein 1 (HMGB1), a protein produced during cellular stress and a putative cause of chronic inflammation [25-27]. To understand the degree to which levels of HMGB1 and RAGE, proteins secreted during cellular stress, associate with sCD14 in PLWH and SIV-infected NHPs, here we measured plasma levels of RAGE, HMGB1, sCD14 and other markers associated with damage to the epithelial barrier of the GI tract such as intestinal fatty acid binding protein (IFABP) in cohorts of ART naïve and ART-treated PLWH and SIV-uninfected and SIV-infected NHPs. We find that neither HMGB1 nor RAGE are likely to be significant causes of elevated sCD14 levels observed routinely in progressively infected PLWH and SIV-infected NHPs.
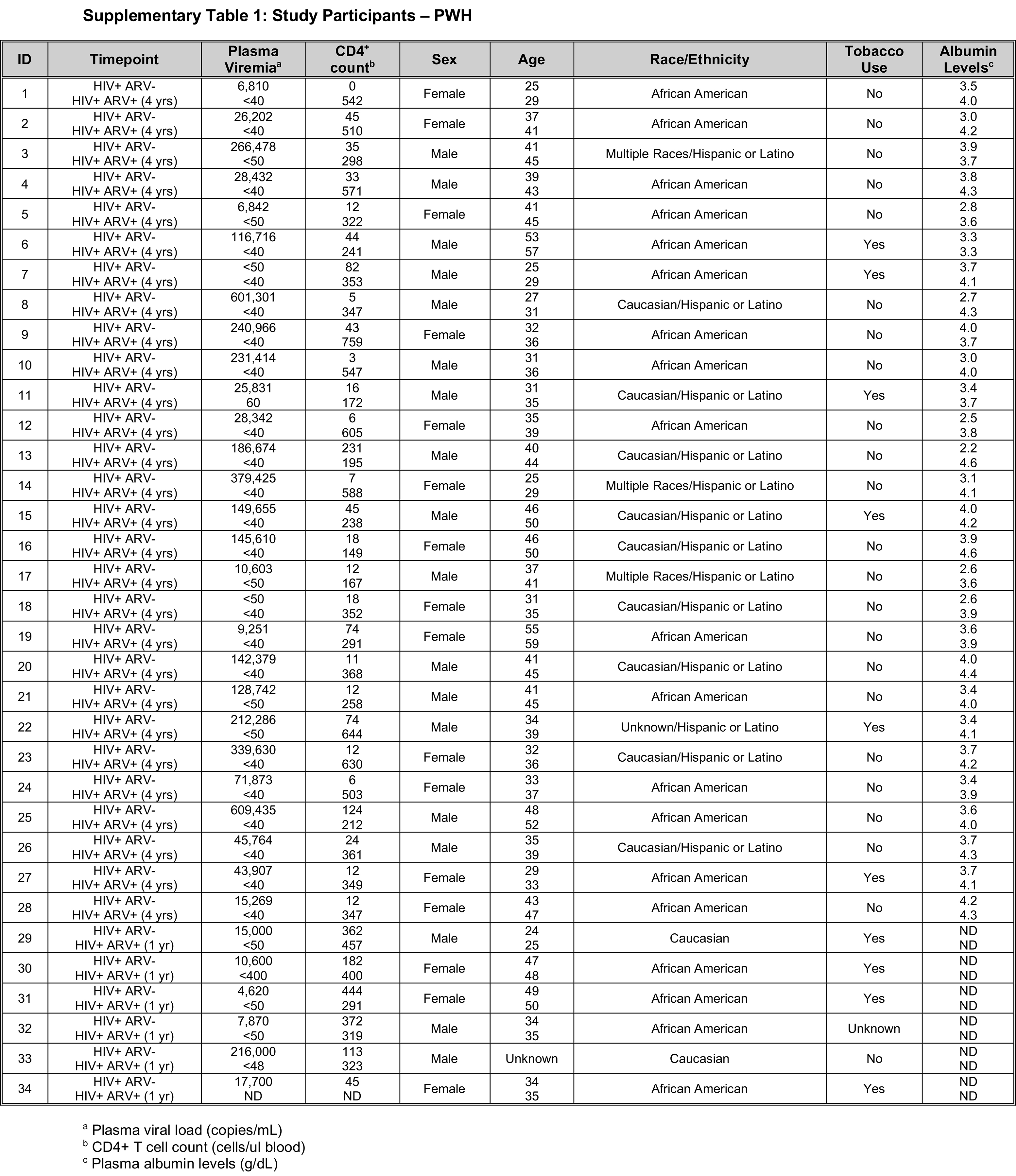
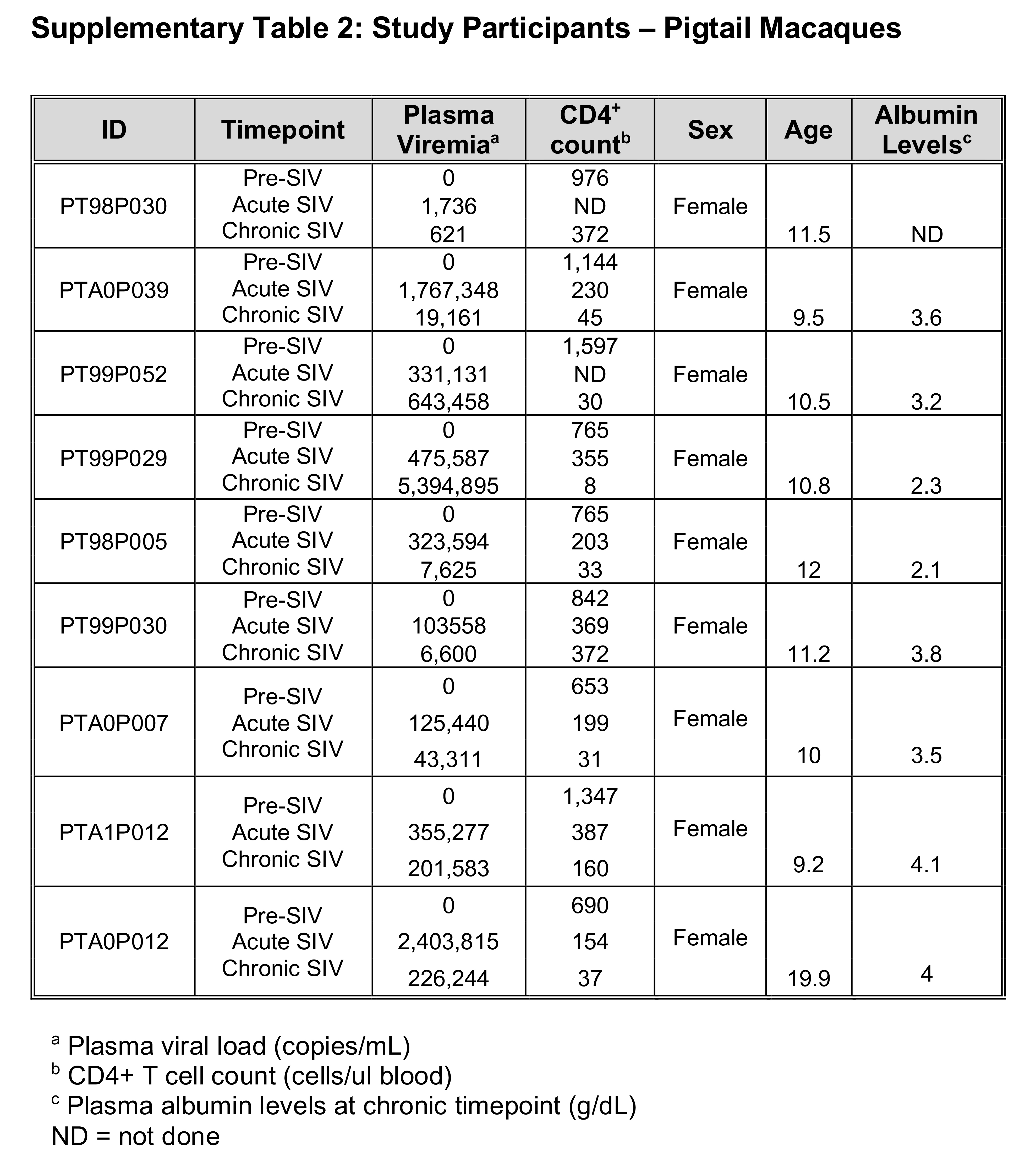
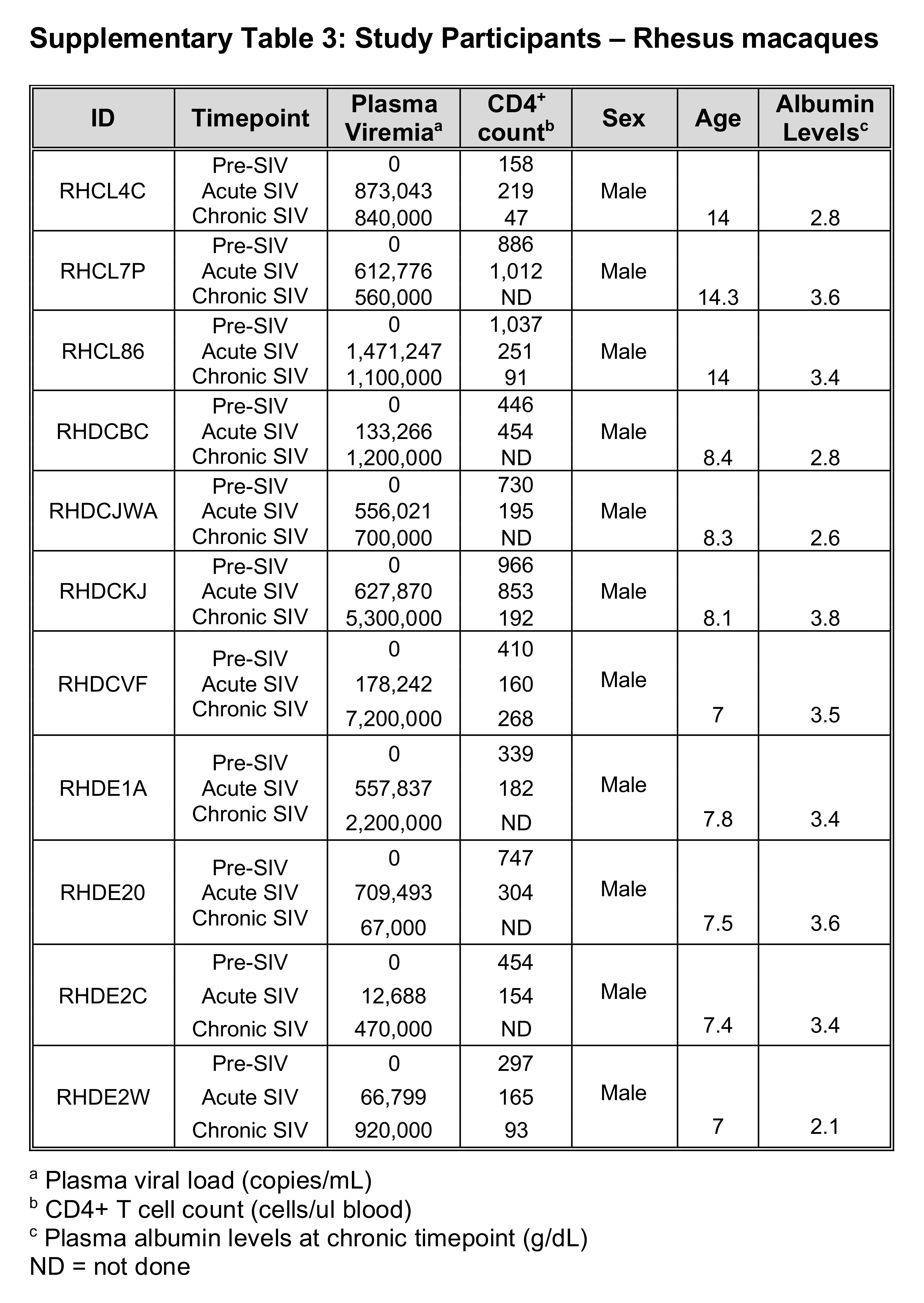
Cohorts for this study consisted of longitudinal samples from 34 PLWH sampled prior to administration of ART and either one or four years after (Supplementary Table 1); 9 pigtail macaque monkeys (PTs) and 11 rhesus macaque monkeys (RMs) followed longitudinally pre-SIV infection and during acute (day 28 or 29 post-SIV infection) and chronic infection (day 197 or later post-SIV infection) (Supplementary Tables 2 and 3). All NHPs in this study were experimentally infected with SIVmac239.
All human study participants provided written informed consent prior to the study under institutional review board-approved protocol: Immune reconstitution syndrome in HIV-infected patients taking antiretroviral therapy (IRIS, NCT00286767) to NIAID, and protocol number 1011003018, Pulmonary CD4 T cell repopulation in immune reconstitution syndrome to the Indiana University.
All NHPs included in this study were housed and cared for in accordance with standards put forth by the American Association for the Accreditation of Laboratory Animal Care (AAALAC). All procedures were performed with approval and in accordance with the Institutional Animal Care and Use Committee within the National Institute of Allergy and Infectious Diseases (animal study protocol LVD26).
Plasma and peripheral blood mononuclear cells (PBMCs) were isolated from whole blood by standard density centrifugation and frozen (plasma) or cryopreserved (PBMC) so that all longitudinal study samples could be analyzed together.
We measured plasma levels of HMGB1, sCD14, RAGE, IFABP, and zonulin by enzyme-linked immunosorbent assay (ELISA) in human and non-human primate samples (Table 1). All markers were analyzed according to the manufacturer’s instructions.
Table 1.
|
|
|
|
|
|
|
HMGB1 |
Human, RM, PT |
Antibodies Online |
#abx151824 |
1:20 |
|
sCD14 |
Human, RM, PT |
R&D Systems |
#DC140 |
1:300 |
|
RAGE |
Human |
R&D Systems |
#DRG00 |
undiluted |
|
RAGE |
RM, PT |
My Biosource |
#MBS746886 |
undiluted |
|
IFABP |
Human |
Hycult |
#HK406-02 |
1:4 |
|
IFABP |
RM, PT |
My Biosource |
#MBS740424 |
undiluted |
|
Zonulin |
Human, RM, PT |
Alpco |
#30-ZONSHU-E01 |
1:20 |
SIV viral RNA levels in plasma were determined by real-time RT-PCR (ABI Prism 7900 sequence detection system; Applied Biosystems) using primer pairs corresponding to SIVmac239 gag gene sequences (forward, nucleotides 1181–1208, and reverse, 1338-1317). HIV viral RNA levels in plasma were determined by either a RTPCR COBAS 1.5 HIV Assay (Roche, with a lower limit of detection of 50 HIV copies/mL) or a RealTime HIV Assay (Abbott, with a lower limit of detection of 40 HIV copies/mL).
CD4+ T-cell counts were calculated from complete blood counts and CD4+ lymphocyte percentages determined via flow cytometric phenotypic profiling of PBMC samples from the same timepoints.
All statistical analyses were performed using Prism v8.0 (GraphPad Software). The Wilcoxon t test (matched pairs, two-way, nonparametric) was used for all analyses between longitudinal samples (Figure 1). All correlational analyses were performed using the Spearman rank test (Figures 2-4). Given correlations were measured in three different primate species, multiple comparison corrections were not considered.

Here we assayed samples from PLWH pre-treatment and following ART (n = 34) and Asian macaque NHPs prior to and throughout SIV-infection (total n = 20, 9 pigtailed macaques, PTs, and 11 rhesus macaques, RMs) (Supplementary Tables 1-3). PLWH had a mean CD4+ count of 75 cells/ml (range 0 to 444 cells/ml) prior to ART and exhibited a significant increase following ART (P < 0.0001, mean count = 385 cells/ml, range 149 to 759 cells/ml). The PLWH cohort had a mean plasma viral load of 127,698 copies (c)/mL (ranging from < 50 (2 patients) to 609,435 c/mL), with all of the study participants having viral loads below or close to the limit of detection after ART of at least 1 year (Supplemental Table 1).
SIV-infected cohorts had a mean acute plasma viral load of 584,338 c/mL (ranging from 1,736 to 2,403,815 c/mL) and mean chronic viral load of 1,267,104 c/mL (ranging from 621 to 7,200,000 c/mL). These SIV-infected cohorts demonstrated a decline in CD4+ T cell counts from pre-SIV infection through acute and chronic SIV-infection (with mean CD4+ T cell counts of 758 cells/ml for pre-, 325 cells/ml acute-, and 127 cells/ml chronic- SIV timepoints) (Supplemental Tables 2-3).
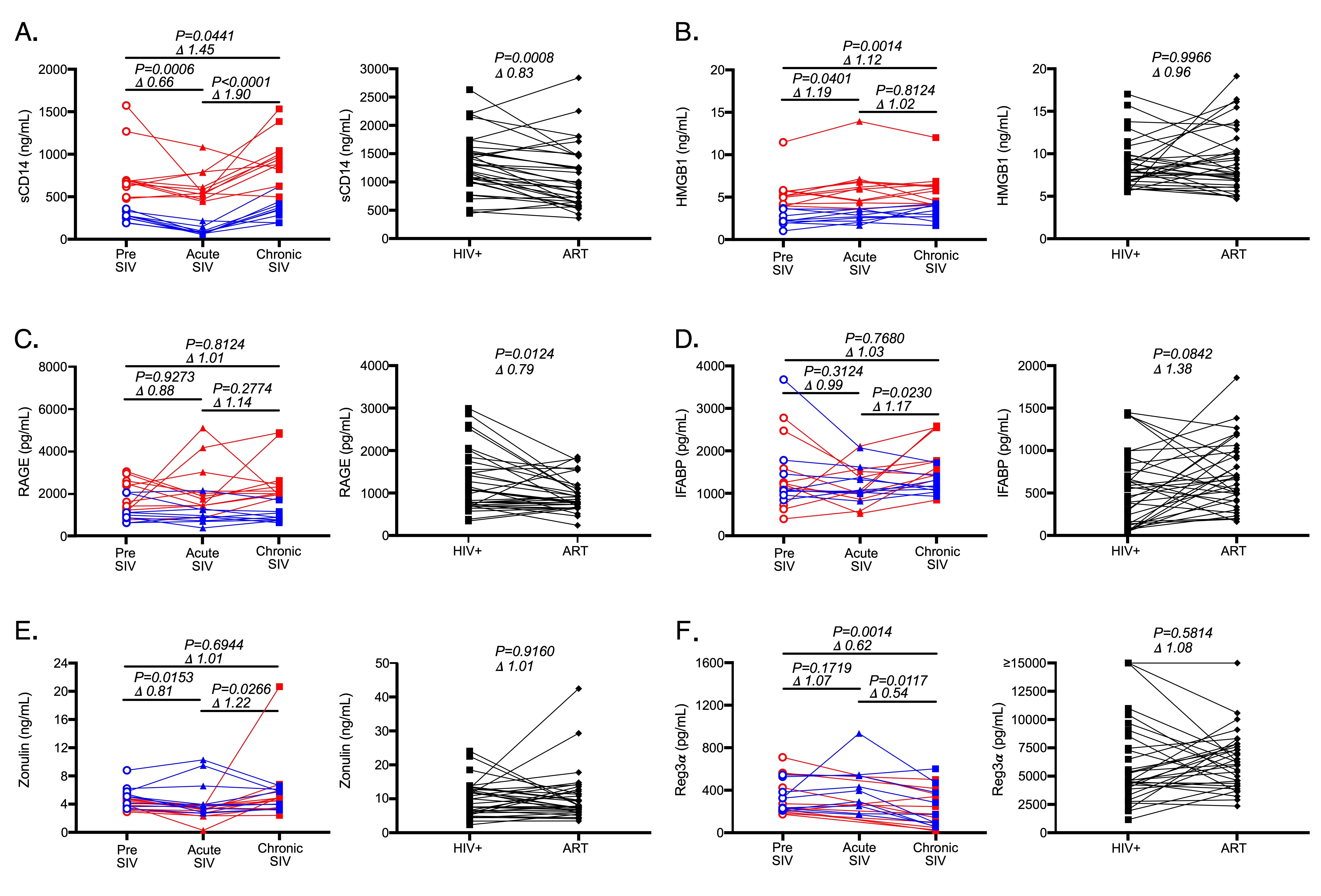
Consistent with several previous studies, we observed a marked increase in plasma sCD14 levels in SIV-infected NHPs from pre-SIV to chronic-SIV and from acute to chronic SIV-infection timepoints (Figure 1A, P = 0.04 and P < 0.0001) and significant decreases in the plasma levels of sCD14 in plasma of PLWH following antiretroviral treatment (ART, P = 0.0008, Figure 1A) [1,3, 15]. Interestingly, sCD14 levels decreased during acute SIV infection (Figure 1A). While we have previously seen these levels to be moderately elevated during acute HIV infection [1], this apparent discrepancy is likely due to the well controlled longtidutinal analysis we were able to perform in NHPs. In order to determine if HMGB1 plasma levels followed similar patterns as sCD14 plasma levels during HIV/SIV infection and following ART, we next assayed this analyte in the same cohorts (Figure 1B). We found that HMGB1 levels were elevated from pre-SIV infection in both acute (P = 0.04) and chronic (P = 0.001) SIV infection timepoints; however, HMGB1 was not increased between acute and chronic SIV infection. In PLWH, plasma levels of HMGB1 demonstrated no significant consistent changes following ART (Figure 1B). Thus, alterations in the levels of sCD14 and HMGB1 were not consistent across SIV infection or after administration of ART in PLWH.
We next assessed levels of RAGE in plasma given its ability to induce sCD14 in vitro and in vivo. In Asian macaques levels of RAGE were entirely unaffected over the course of SIV infection (Figure 1C). In PLWH, RAGE levels significantly decreased after administration of ART (P = 0.01, Figure 1C). Thus, neither HMGB1 nor RAGE levels mirrored sCD14 level alterations which occur over the course of SIV infection of Asian macaques. However, RAGE levels in PLWH did follow a decreasing trend similar to that seen for sCD14 plasma levels after administration of ART in PLWH (P = 0.01, Figure 1C).
Microbial translocation occurs when there is a loss of/or damage to the GI tract epithelial barrier. Epithelial damage is thought to result in rapid release of epithelial fatty acid binding proteins and extracellular proteins involved in epithelial tight junctions into circulation [28, 29]. To assess the reliability of proteins released during epithelial damage as potential markers of microbial translocation, we measured plasma levels of IFABP and zonulin, a protein that functions to maintain intercellular junctions, in plasma. As Asian macaques progressed from acute to chronic infection, both IFABP (P = 0.02, Figure 1D) and zonulin (P =0 .03, Figure 1E) increased. In PLWH, 1-4 years of ART treatment was insufficient to result in significant decreases in these markers of intestinal damage (Figure 1D-E) consistent with previous reports [30]. In fact, IFABP levels actually tended to increase (P = 0.08, Figure 1D), likely due to increased epithelial cell turnover to repair the barrier. These data suggest that prolonged ART does not completely reduce GI tract damage observed in PLWH. Like HMGB1 and RAGE, these results did not track with sCD14 dynamics.
Another systemic biomarker which has recently been suggested to be associated with microbial translocation is Regenerating islet-derived protein 3a (Reg3a), an antimicrobial peptide produced by the GI tract epithelium [31]. Plasma levels of Reg3a have been shown to be elevated in chronic HIV infection and to correlate with plasma levels of lipopolysaccharide, pro-inflammatory cytokines, and percentages of activated CD4+ and CD8+ T cells [32]. Surprisingly, in our cohort of Asian macaques, plasma levels of Reg3a were unchanged during acute infection as compared to pre-infection levels, and actually decreased into chronic infection (P = 0.01, Figure 1F). Moreover, we found no significant differences in circulating plasma levels of Reg3a in our human cohort following ART (Figure 1F), while previous reports suggested these levels decrease in plasma of PLWH treated with ART [32].
Although markers of cellular stress, HMGB1 and RAGE, did not mirror sCD14 dynamics over the course of SIV infection or following ART in PLWH, we sought to determine if either analyte correlated with sCD14 or systemic levels of gut barrier damage. Positive associations would be supportive of their direct role in production of sCD14 in vivo. Even though HMGB1 levels were not decreased in PLWH treated with ART, changes over time in sCD14 trended towards a positive correlation with HMGB1 (human results in black r = 0.3303, P = 0.0564, Figure 2A). This association was not observed in either NHP species (RM results shown in red; PT results shown in blue Figure 2A). Interestingly, although longitudinal analysis of human plasma levels of RAGE trended consistently with sCD14 systemic plasma levels (Figure 1A and 1C), there were no significant correlations between changes in RAGE plasma level changes and longitudinal changes in sCD14 levels in either humans or NHPs (Figure 2B). sCD14 levels were not associated with IFABP or zonulin levels in any primate species (Figure 2C and 2D).
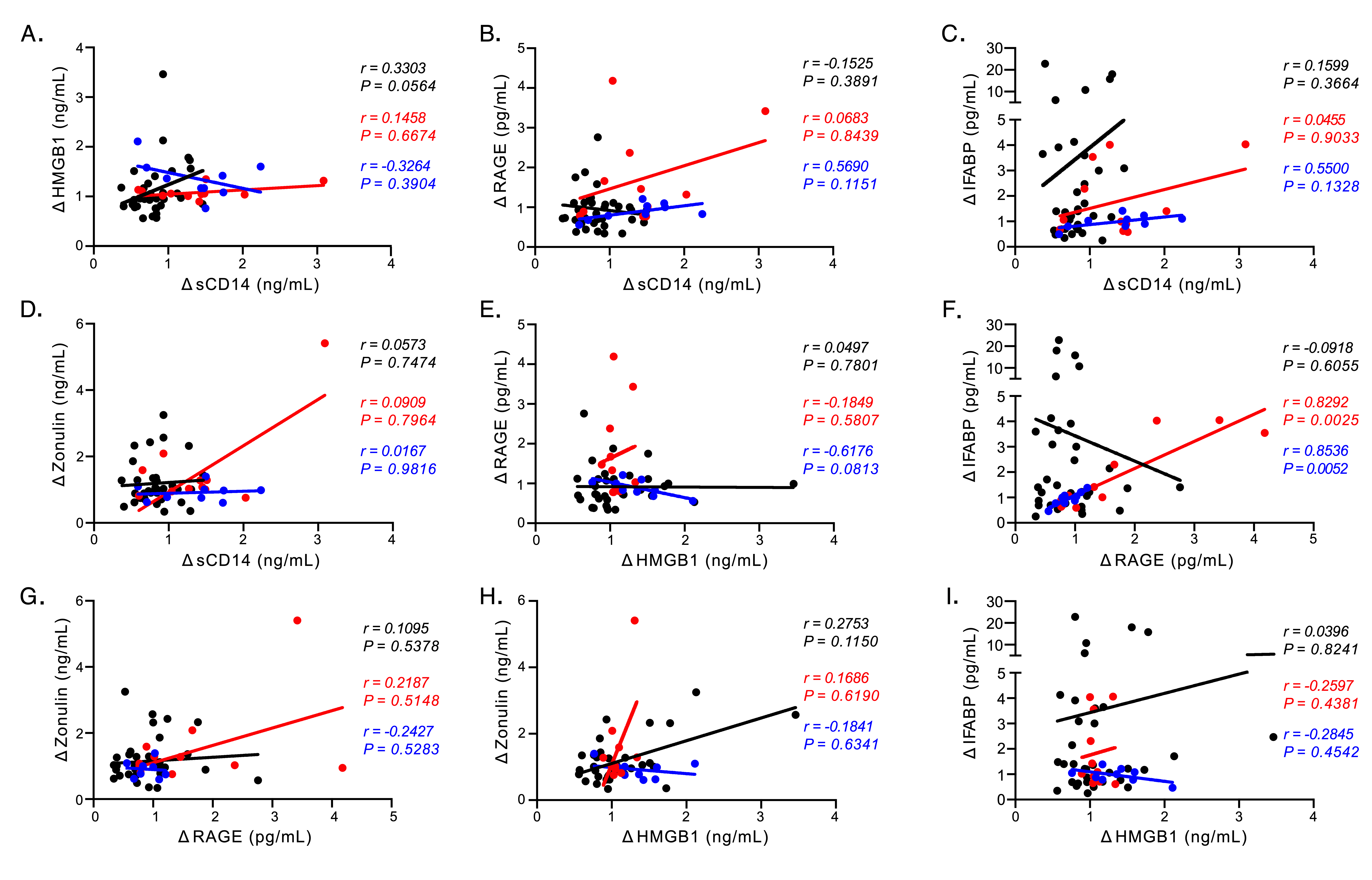
Figure 2. Plasma biomarker associations. (A-I) Correlational analyses between longitudinal changes in plasma biomarkers in humans (black), RMs (red), and PTs (blue). All associations were analyzed using the Spearman rank test.
We next extended this analysis to determine if changes in HMGB1 or RAGE were associated with markers of GI tract damage. While HMGB1 and RAGE can both be elevated as a result of cellular damage, these were not associated with one another in any of the primate species we studied (Figure 2E). We did find strong associations between changes in plasma levels of RAGE and IFABP in both NHP groups (RM r = 0.83 and P = 0.0025; PT r = 0.85 and P = 0.0052, Figure 2F) with no such association being observed in our human cohort. However, changes in RAGE did not associate with change in zonulin levels in either NHP cohort (Figure 2G). Furthermore, changes in HMGB1 plasma levels did not correlate with either of the markers of epithelial damage we assayed (zonulin Figure 2H and IFABP Figure 2I). From these analyses, the data suggest that in progressive HIV/SIV infections neither HMGB1 nor RAGE contribute to elevated levels of biomarkers used as surrogates for microbial translocation.
We next determined whether canonical measures of HIV/SIV disease progression were associated with levels of any plasma biomarkers we studied. In particular, we studied associations with changes in CD4+ T cell counts (Figure 3) and changes in plasma viral load (Figure 4) over time. For PLWH we looked at the change in value from chronic infection to post-ART (D ART/chronic baseline), while for NHPs we looked at the change in values from pre- to chronic SIV-infection (D chronic/pre). Among the five studied biomarkers, only HMGB1 (Figure 3B) and RAGE (Figure 3C) showed correlations with changes in the numbers of CD4+ T cells in peripheral blood; however these associations were seen only in the PT cohort- no other correlations were observed.
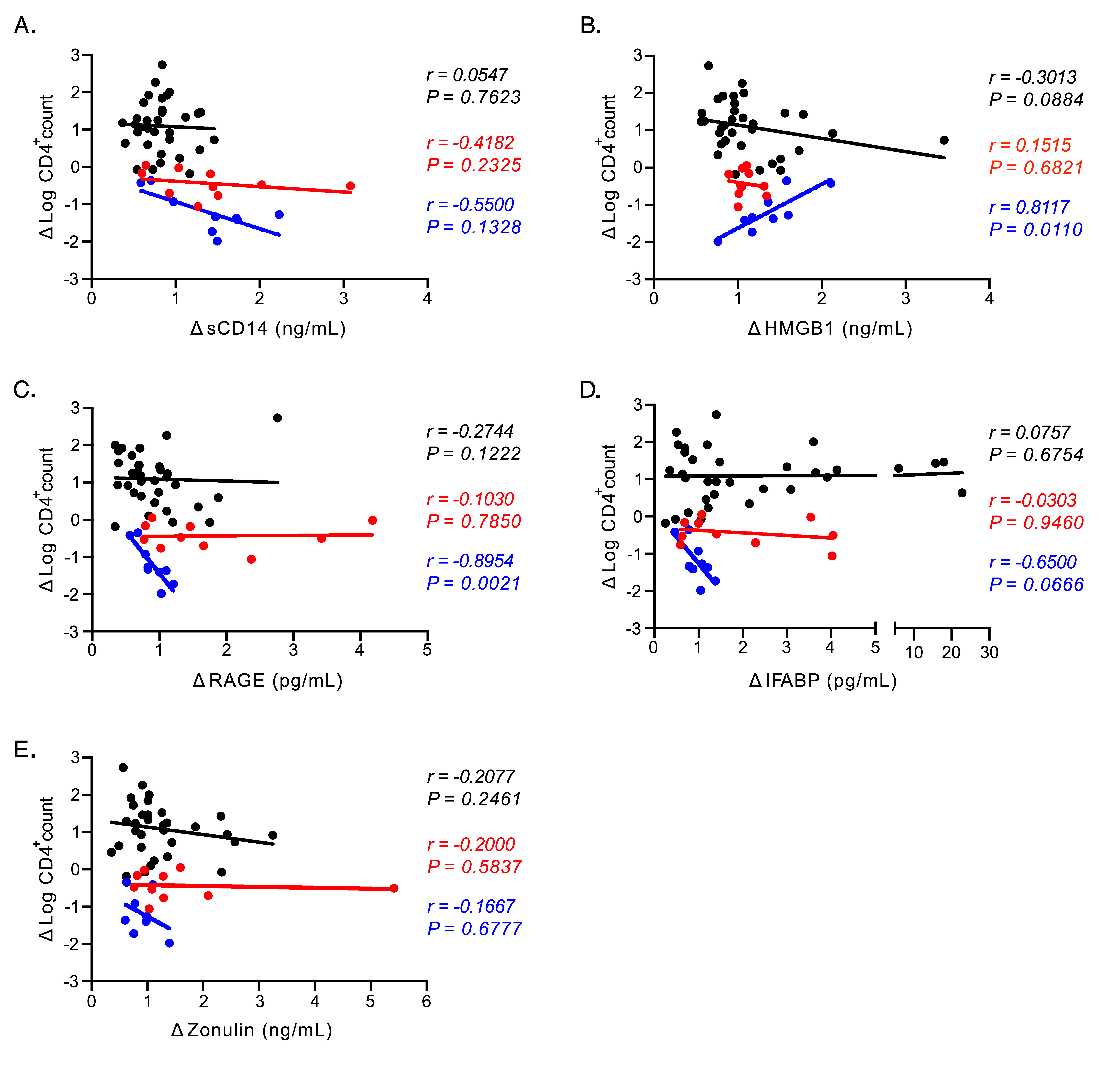
Figure 3. Plasma biomarker associations with CD4 T cell frequencies. (A-E) Correlational analyses between changes in CD4+ T cell numbers in peripheral blood and changes in plasma biomarkers in humans (black), RMs (red), and PTs (blue) for (A) sCD14, (B) HMGB1, (C) RAGE), (D) IFABP, and (E) Zonulin. All associations were analyzed using the Spearman rank test.
Lastly, direct or indirect effects of viral components (such as viral RNA) can be directly stimulatory to the immune system, thus alternately explaining elevated levels of sCD14 and other biomarkers used as surrogates for microbial translocation [33-35]. Therefore, we measured relationships between plasma viremia and each of the biomarkers we studied during chronic infection (Figure 4). In PTs HMGB1 was actually negatively correlated with plasma viremia (Figure 4B), while in humans RAGE levels positively correlated (Figure 4C). However, these associations were not observed across all three primate species and we found no associations between plasma viremia and either sCD14 (Figure 4A) or IFABP (Figure 4D). These data argue against plasma viremia (either directly or indirectly) leading to altered levels of these biomarkers.
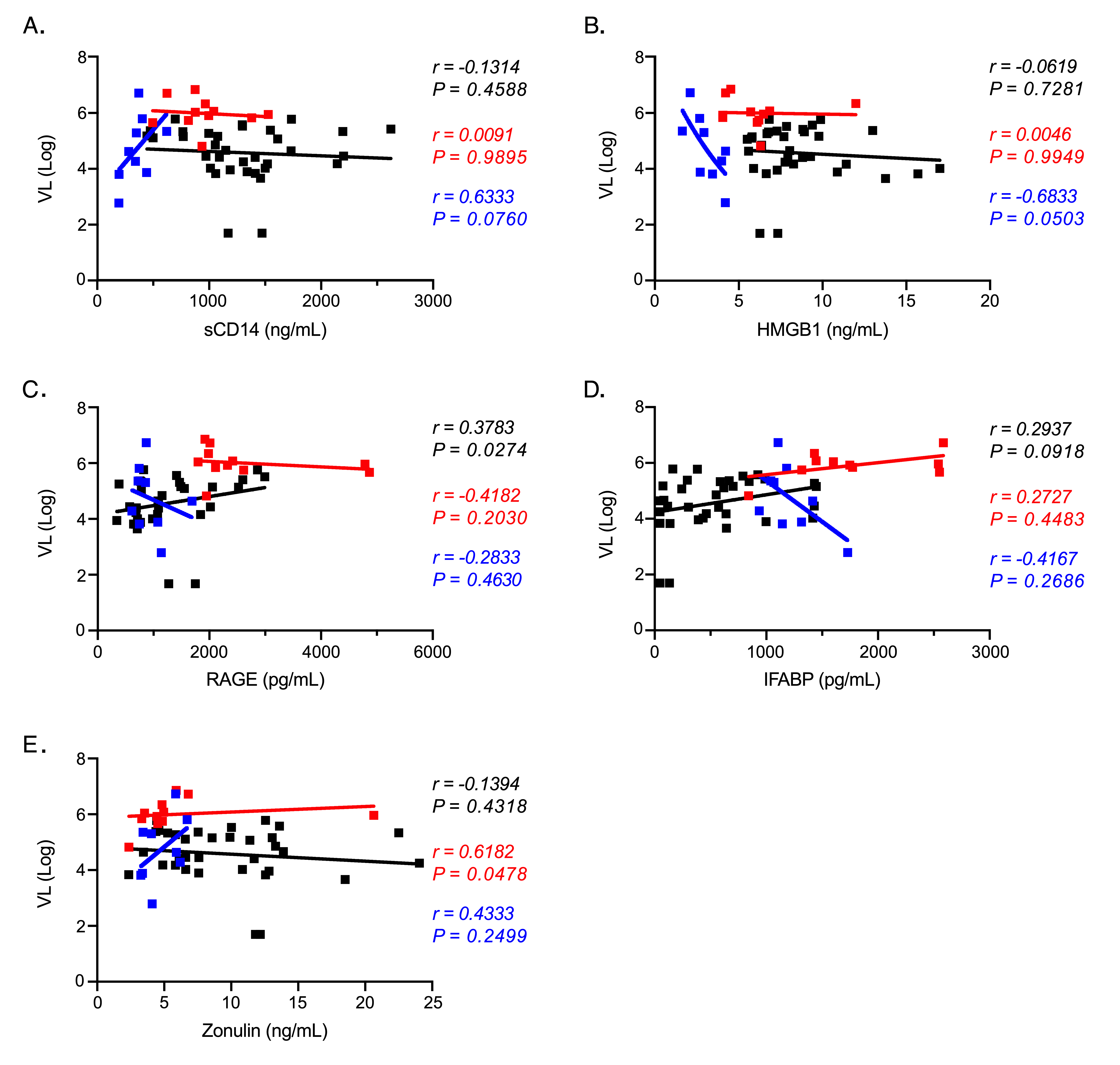
Figure 4. Plasma biomarker associations with plasma viral load. (A-E) Correlational analyses between SIV or HIV plasma viral load during chronic infection and plasma biomarkers in humans (black), RMs (red), and PTs (blue) for (A) sCD14, (B) HMGB1, (C) RAGE), (D) IFABP, and (E) Zonulin. All associations were analyzed using the Spearman rank test.
Microbial translocation is a well described phenomenon in chronically infected PLWH and SIV-infected Asian macaques [1,3, 20]. Even after decades of ART - particularly if ART was initiated when nadir CD4+ T cell counts in peripheral blood were low - the damage that had occurred to the GI tract is not completely reversed, (30,36, 37]. Residual inflammation which occurs in these treated individuals leads to increased mortality from different malignancies and cardiovascular disease and may be attributed, at least partially, to ongoing microbial translocation [11,30,38, 39]. While the current World Health Organization’s recommendations are to initiate therapy as early as possible in all PLWH [40], most PLWH currently taking ART did not initiate therapy until the chronic phase of infection. These individuals may benefit from supplemental therapies which restore the GI tract and reduce microbial translocation and inflammation. Thus, there is a great need to identify putative biomarkers which can be used to monitor efficacy of strategies aimed at reducing microbial translocation and inflammation. Of the numerous plasma analytes which might fulfill this purpose, sCD14 is, by far, most frequently studied. Although sCD14 levels represent bioactivity of microbial products, several studies have shown that sCD14 levels can be elevated in the apparent absence of microbial translocation [22, 24]. While CD14 is, indeed, a specific receptor for LPS, HMGB1 is also a ligand for CD14 [25-27].
Here we sought to determine how levels of particular plasma biomarkers associated with microbial translocation and inflammation change over the course of SIV infection of NHPs and after administration of ARTs in HIV-1-infected individuals. We also sought to determine whether HMGB1 and RAGE might contribute to the elevated plasma levels of sCD14 observed in progressive HIV/SIV infections. Our data suggest that HMGB1 and RAGE are not associated with elevated levels of sCD14 observed in PLWH and SIV-infected NHPs given that neither HMGB1 nor RAGE levels reliably changed over time as NHPs became SIV-infected or PLWH initiated ARTs. Thus, elevated sCD14 is unlikely to be attributed to cellular death in these cases. We also do not find evidence that markers of intestinal epithelial damage correlate with sCD14, suggesting that these epithelial markers themselves are not sufficient biomarkers of microbial translocation and epithelial turnover after administration of ART could contribute to elevated levels of IFABP.
There is significant interest in understanding the mechanisms underlying microbial translocation, the degree to which microbial translocation contributes to inflammation observed in PLWH, and in developing therapeutic interventions which ameliorate microbial translocation [3, 41-43]. Assays to detect bacterial products in plasma lack sensitivity and reproducibility and assays to measure bacterial products in tissue require invasive procedures. While our study included a small cohort of NHPs and PLWH, and would have benefitted from inclusion of tissue biopsies, we believe our study demonstrates that measurement of sCD14 in plasma in conjunction with proteins which are released into circulation when GI tract epithelial cells are damaged (IFABP and zonulin) are currently the best available surrogates of systemic microbial translocation. We also believe that tissue measurement of microbial products remains the most robust measurement of microbial translocation. As next generation sequencing approaches become more available and accessible, the ability to detect nucleic acids corresponding to microbes in circulation may alleviate these issues [44].
Funding for this study was provided in part by the Division of Intramural Research/NIAID/NIH. The content of this publication does not necessarily reflect the views or policies of DHHS, nor does the mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government. KK was funded by R01 HL083468.
The authors declare no financial conflicts of interest.
1. Brenchley JM, Price DA, Schacker TW, Asher TE, Silvestri G, Rao S, Kazzaz Z, Bornstein E, Lambotte O, Altmann D, Blazar BR, Rodriguez B, Teixeira-Johnson L, Landay A, Martin JN, Hecht FM, Picker LJ, Lederman MM, Deeks SG, Douek DC. Microbial translocation is a cause of systemic immune activation in chronic HIV infection. Nat Med. 2006;12(12):1365-71. PubMed PMID. doi: 10.1038/nm1511
2. Estes JD, Harris LD, Klatt NR, Tabb B, Pittaluga S, Paiardini M, Barclay GR, Smedley J, Pung R, Oliveira KM, Hirsch VM, Silvestri G, Douek DC, Miller CJ, Haase AT, Lifson J, Brenchley JM. Damaged intestinal epithelial integrity linked to microbial translocation in pathogenic simian immunodeficiency virus infections. PLoS Pathog. 2010;6(8):e1001052. PubMed PMID: 2924359. doi: 10.1371/journal.ppat.1001052
3. Klatt NR, Funderburg NT, Brenchley JM. Microbial translocation, immune activation, and HIV disease. Trends Microbiol. 2013;21(1):6-13. PubMed PMID: 3534808. doi: 10.1016/j.tim.2012.09.001
4. Brenchley JM, Schacker TW, Ruff LE, Price DA, Taylor JH, Beilman GJ, Nguyen PL, Khoruts A, Larson M, Haase AT, Douek DC. CD4+ T cell depletion during all stages of HIV disease occurs predominantly in the gastrointestinal tract. J Exp Med. 2004;200(6):749-59. PubMed PMID: 2211962. doi: 10.1084/jem.20040874
5. Klatt NR, Estes JD, Sun X, Ortiz AM, Barber JS, Harris LD, Cervasi B, Yokomizo LK, Pan L, Vinton CL, Tabb B, Canary LA, Dang Q, Hirsch VM, Alter G, Belkaid Y, Lifson JD, Silvestri G, Milner JD, Paiardini M, Haddad EK, Brenchley JM. Loss of mucosal CD103+ DCs and IL-17+ and IL-22+ lymphocytes is associated with mucosal damage in SIV infection. Mucosal Immunol. 2012;5(6):646-57. PubMed PMID: 3443541. doi: 10.1038/mi.2012.38
6. Mudd JC, Busman-Sahay K, DiNapoli SR, Lai S, Sheik V, Lisco A, Deleage C, Richardson B, Palesch DJ, Paiardini M, Cameron M, Sereti I, Reeves RK, Estes JD, Brenchley JM. Hallmarks of primate lentiviral immunodeficiency infection recapitulate loss of innate lymphoid cells. Nat Commun. 2018;9(1):3967. PubMed PMID. doi: 10.1038/s41467-018-05528-3
7. Brenchley JM, Paiardini M, Knox KS, Asher AI, Cervasi B, Asher TE, Scheinberg P, Price DA, Hage CA, Kholi LM, Khoruts A, Frank I, Else J, Schacker T, Silvestri G, Douek DC. Differential Th17 CD4 T-cell depletion in pathogenic and nonpathogenic lentiviral infections. Blood. 2008;112(7):2826-35. PubMed PMID. doi: blood-2008-05-159301
8. Swan ZD, Wonderlich ER, Barratt-Boyes SM. Macrophage accumulation in gut mucosa differentiates AIDS from chronic SIV infection in rhesus macaques. Eur J Immunol. 2016;46(2):446-54. PubMed PMID: PMC5751443. doi: 10.1002/eji.201545738
9. Sankaran S, George MD, Reay E, Guadalupe M, Flamm J, Prindiville T, Dandekar S. Rapid onset of intestinal epithelial barrier dysfunction in primary human immunodeficiency virus infection is driven by an imbalance between immune response and mucosal repair and regeneration. J Virol. 2008;82(1):538-45. PubMed PMID: 2224350. doi: JVI.01449-07
10. Li Q, Estes JD, Duan L, Jessurun J, Pambuccian S, Forster C, Wietgrefe S, Zupancic M, Schacker T, Reilly C, Carlis JV, Haase AT. Simian immunodeficiency virus-induced intestinal cell apoptosis is the underlying mechanism of the regenerative enteropathy of early infection. J Infect Dis. 2008;197(3):420-9. PubMed PMID. doi: 10.1086/525046
11. Hsue PY, Waters DD. HIV infection and coronary heart disease: mechanisms and management. Nat Rev Cardiol. 2019. PubMed PMID. doi: 10.1038/s41569-019-0219-9
12. Lohse N, Hansen AB, Pedersen G, Kronborg G, Gerstoft J, Sorensen HT, Vaeth M, Obel N. Survival of persons with and without HIV infection in Denmark, 1995-2005. Ann Intern Med. 2007;146(2):87-95.
13. Lohse N, Obel N. Update of Survival for Persons With HIV Infection in Denmark. Ann Intern Med. 2016;165(10):749-50. PubMed PMID. doi: 10.7326/L16-0091
14. Hileman CO, Funderburg NT. Inflammation, Immune Activation, and Antiretroviral Therapy in HIV. Curr HIV/AIDS Rep. 2017;14(3):93-100. PubMed PMID: PMC5514315. doi: 10.1007/s11904-017-0356-x
15. Jiang W, Lederman MM, Hunt P, Sieg SF, Haley K, Rodriguez B, Landay A, Martin J, Sinclair E, Asher AI, Deeks SG, Douek DC, Brenchley JM. Plasma levels of bacterial DNA correlate with immune activation and the magnitude of immune restoration in persons with antiretroviral-Treated HIV infection. J Infect Dis. 2009;199(8):1177-85. PubMed PMID. doi: 10.1086/597476
16. Garcia-Alvarez M, Berenguer J, Jimenez-Sousa MA, Vazquez-Moron S, Carrero A, Gutierrez-Rivas M, Aldamiz-Echevarria T, Lopez JC, Garcia-Broncano P, Resino S. Optimal vitamin D plasma levels are associated with lower bacterial DNA translocation in HIV/hepatitis c virus coinfected patients. AIDS. 2016;30(7):1069-74. doi: 10.1097/QAD.0000000000001007
17. Pilakka-Kanthikeel S, Kris A, Selvaraj A, Swaminathan S, Pahwa S. Immune activation is associated with increased gut microbial translocation in treatment-naive, HIV-infected children in a resource-limited setting. J Acquir Immune Defic Syndr. 2014;66(1):16-24. PubMed PMID: PMC4386670. doi: 10.1097/QAI.0000000000000096
18. Napirei M, Ricken A, Eulitz D, Knoop H, Mannherz HG. Expression pattern of the deoxyribonuclease 1 gene: lessons from the Dnase1 knockout mouse. Biochem J. 2004;380(Pt 3):929-37. PubMed PMID: PMC1224217. doi: 10.1042/BJ20040046
19. Yao Z, Mates JM, Cheplowitz AM, Hammer LP, Maiseyeu A, Phillips GS, Wewers MD, Rajaram MV, Robinson JM, Anderson CL, Ganesan LP. Blood-Borne Lipopolysaccharide Is Rapidly Eliminated by Liver Sinusoidal Endothelial Cells via High-Density Lipoprotein. J Immunol. 2016;197(6):2390-9. PubMed PMID: PMC5010928. doi: 10.4049/jimmunol.1600702
20. Brenchley JM. Mucosal immunity in human and simian immunodeficiency lentivirus infections. Mucosal Immunol. 2013;6(4):657-65. PubMed PMID. doi: 10.1038/mi.2013.15
21. Kitchens RL, Thompson PA. Modulatory effects of sCD14 and LBP on LPS-host cell interactions. J Endotoxin Res. 2005;11(4):225-9.
22. Shive CL, Jiang W, Anthony DD, Lederman MM. Soluble CD14 is a nonspecific marker of monocyte activation. AIDS. 2015;29(10):1263-5. PubMed PMID: PMC4452959. doi: 10.1097/QAD.0000000000000735
23. Rajamanickam A, Munisankar S, Bhootra Y, Dolla C, Nutman TB, Babu S. Microbial Translocation Associated with an Acute-Phase Response and Elevations in MMP-1, HO-1, and Proinflammatory Cytokines in Strongyloides stercoralis Infection. Infect Immun. 2017;85(1). PubMed PMID: PMC5203642. doi: 10.1128/IAI.00772-16
24. De Voeght A, Maes N, Moutschen M. sCD14 is not a bona-fide biomarker of microbial translocation in HIV-1-infected Africans living in Belgium. AIDS. 2016;30(6):921-4. doi: 10.1097/QAD.0000000000000996
25. Ibrahim ZA, Armour CL, Phipps S, Sukkar MB. RAGE and TLRs: relatives, friends or neighbours? Mol Immunol. 2013;56(4):739-44. doi: 10.1016/j.molimm.2013.07.008
26. Wang H, Ward MF, Fan XG, Sama AE, Li W. Potential role of high mobility group box 1 in viral infectious diseases. Viral Immunol. 2006;19(1):3-9. PubMed PMID: PMC1782047. doi: 10.1089/vim.2006.19.3
27. Li W, Sama AE, Wang H. Role of HMGB1 in cardiovascular diseases. Curr Opin Pharmacol. 2006;6(2):130-5. PubMed PMID: PMC1782046. doi: 10.1016/j.coph.2005.10.010
28. Cummins G, Yung DE, Cox BF, Koulaouzidis A, Desmulliez MPY, Cochran S. Luminally expressed gastrointestinal biomarkers. Expert Rev Gastroenterol Hepatol. 2017;11(12):1119-34. PubMed PMID. doi: 10.1080/17474124.2017.1373017
29. Demir E, Ozkan H, Seckin KD, Sahtiyanci B, Demir B, Tabak O, Kumbasar A, Uzun H. Plasma Zonulin Levels as a Non-Invasive Biomarker of Intestinal Permeability in Women with Gestational Diabetes Mellitus. Biomolecules. 2019;9(1). PubMed PMID: PMC6359541. doi: 10.3390/biom9010024
30. Sandler NG, Wand H, Roque A, Law M, Nason MC, Nixon DE, Pedersen C, Ruxrungtham K, Lewin SR, Emery S, Neaton JD, Brenchley JM, Deeks SG, Sereti I, Douek DC, Group ISS. Plasma levels of soluble CD14 independently predict mortality in HIV infection. J Infect Dis. 2011;203(6):780-90. PubMed PMID: 3071127. doi: 10.1093/infdis/jiq118
31. Marafini I, Di Sabatino A, Zorzi F, Monteleone I, Sedda S, Cupi ML, Antenucci C, Biancheri P, Giuffrida P, Di Stefano M, Corazza GR, Pallone F, Monteleone G. Serum regenerating islet-derived 3-alpha is a biomarker of mucosal enteropathies. Aliment Pharmacol Ther. 2014;40(8):974-81. doi: 10.1111/apt.12920
32. Isnard S, Ramendra R, Dupuy FP, Lin J, Fombuena B, Kokinov N, Kema I, Jenabian MA, Lebouche B, Costiniuk CT, Ancuta P, Bernard NF, Silverman MS, Lakatos PL, Durand M, Tremblay C, Routy JP, Montreal Primary Hiv Infection Study tCCoHIVSP, the Canadian HIV, Aging Cohort G. Plasma Levels of C-Type Lectin REG3alpha and Gut Damage in People With Human Immunodeficiency Virus. J Infect Dis. 2020;221(1):110-21. PubMed PMID: PMC6910878. doi: 10.1093/infdis/jiz423
33. Alter G, Suscovich TJ, Teigen N, Meier A, Streeck H, Brander C, Altfeld M. Single-stranded RNA derived from HIV-1 serves as a potent activator of NK cells. J Immunol. 2007;178(12):7658-66.
34. Meier A, Chang JJ, Chan ES, Pollard RB, Sidhu HK, Kulkarni S, Wen
TF, Lindsay RJ, Orellana L, Mildvan D, Bazner S, Streeck H, Alter G,
Lifson JD, Carrington M, Bosch RJ, Robbins GK, Altfeld M. Sex
differences in the Toll-like receptor-mediated response of plasmacytoid
dendritic cells to HIV-1. Nat Med. 2009;15(8):955-9. doi:
nm.2004
35. Caron G, Duluc D, Fremaux I, Jeannin P, David C, Gascan H, Delneste
Y. Direct stimulation of human T cells via TLR5 and TLR7/8: flagellin
and R-848 up-regulate proliferation and IFN-gamma production by memory
CD4+ T cells. J Immunol. 2005;175(3):1551-7.
36. Baroncelli S, Galluzzo CM, Pirillo MF, Mancini MG, Weimer LE,
Andreotti M, Amici R, Vella S, Giuliano M, Palmisano L. Microbial
translocation is associated with residual viral replication in
HAART-treated HIV+ subjects with <50copies/ml HIV-1 RNA. J Clin
Virol. 2009;46(4):367-70. doi: 10.1016/j.jcv.2009.09.011
37. Chege D, Sheth PM, Kain T, Kim CJ, Kovacs C, Loutfy M, Halpenny R,
Kandel G, Chun TW, Ostrowski M, Kaul R. Sigmoid Th17 populations, the
HIV latent reservoir, and microbial translocation in men on long-term
antiretroviral therapy. AIDS. 2011;25(6):741-9. PubMed PMID. doi: 10.1097/QAD.0b013e328344cefb
38. Alcaide ML, Parmigiani A, Pallikkuth S, Roach M, Freguja R, Della
Negra M, Bolivar H, Fischl MA, Pahwa S. Immune activation in
HIV-infected aging women on antiretrovirals--implications for
age-associated comorbidities: a cross-sectional pilot study. PLoS One.
2013;8(5):e63804. PubMed PMID: 3665816. doi:
10.1371/journal.pone.0063804
39. Palella FJ, Jr., Baker RK, Moorman AC, Chmiel JS, Wood KC, Brooks
JT, Holmberg SD, Investigators HIVOS. Mortality in the highly active
antiretroviral therapy era: changing causes of death and disease in the
HIV outpatient study. J Acquir Immune Defic Syndr. 2006;43(1):27-34.
doi: 10.1097/01.qai.0000233310.90484.16
40. Guidelines for Managing Advanced HIV Disease and Rapid Initiation
of Antiretroviral Therapy. WHO Guidelines Approved by the Guidelines
Review Committee. Geneva2017.
41. Brenchley JM, Douek DC. Microbial translocation across the GI
tract. Annu Rev Immunol. 2012;30:149-73. doi: 10.1146/annurev-immunol-020711-075001
42. Hunt PW. HIV and inflammation: mechanisms and consequences. Curr
HIV/AIDS Rep. 2012;9(2):139-47. doi:
10.1007/s11904-012-0118-8
43. Tincati C, Douek DC, Marchetti G. Gut barrier structure, mucosal
immunity and intestinal microbiota in the pathogenesis and treatment of
HIV infection. AIDS Res Ther. 2016;13:19. PubMed PMID: 4828806. doi:
10.1186/s12981-016-0103-1
44. Ericsen AJ, Lauck M, Mohns MS, DiNapoli SR, Mutschler JP, Greene
JM, Weinfurter JT, Lehrer-Brey G, Prall TM, Gieger SM, Buechler CR,
Crosno KA, Peterson EJ, Reynolds MR, Wiseman RW, Burwitz BJ, Estes JD,
Sacha JB, Friedrich TC, Brenchley JM, O'Connor DH. Microbial
Translocation and Inflammation Occur in Hyperacute Immunodeficiency
Virus Infection and Compromise Host Control of Virus Replication. PLoS
Pathog. 2016;12(12):e1006048. PubMed PMID: 5142784. doi:
10.1371/journal.ppat.1006048
Footnotes
Submitted March 2, 2020 | Accepted April 9, 2020 | Published April 24, 2020
Supplementary Material