Mark E. Obrenovich1,2,3,4*, George E. Jaskiw5,6, Renliang Zhang7, Belinda Willard7, Curtis J. Donskey 6,8
1
Pathology and Laboratory Medicine Service, Louis Stokes Cleveland
Department of Veterans Affairs Medical Center, Cleveland, Ohio
2
Research Service, Louis Stokes Cleveland Department of Veterans Affairs
Medical Center, Cleveland, Ohio
3
Department of Chemistry, Case Western Reserve University, Cleveland, Ohio
4
Department of Medicinal and Biological Chemistry, College of Pharmacy and
Pharmaceutical Sciences, University of Toledo, Toledo, Ohio
5
Psychiatry Service, Louis Stokes Cleveland Department of Veteran’s Affairs
Medical Center, Cleveland, Ohio
6
School of Medicine, Case Western Reserve University, Cleveland, Ohio
7Proteomics and Metabolomics Core, Lerner Research Institute, Cleveland
Clinic Foundation, Cleveland, Ohio
8
Geriatric Research, Education and Clinical Center, Louis Stokes Cleveland
Department of Veterans Affairs Medical Center, Cleveland, Ohio
George E. Jaskiw
Psychiatry Service 116(A)
Louis Stokes Cleveland DVAMC
10701 East Blvd.
Cleveland, OH 44106
(216) 791-3800
gxj5@case.edu
Background: Urinary levels of small molecules generated by the gut microbiota (GMB) constitute potential biomarkers for the state of the GMB. Such metabolites include numerous small phenolic molecules linked to anaerobic bacteria, particularly Clostridium species. Due to multiple technical challenges, however, the relationship between these chemicals and the GMB remains poorly characterized. Improved, high-performance liquid chromatography-mass spectrometry (LC-MS)-based metabolomic analysis can now reliably separate and quantify low levels of multiple small phenolic molecules and their structural isomers.
Methods: CF-1 (female mice) were treated over 2 consecutive days with either i) vehicle, ii) one of 2 different antibiotic regimens (clindamycin or piperacillin/tazobactam) known to inhibit intestinal anaerobes and promote colonization by Clostridium difficile and other pathogens or iii) an antibiotic (aztreonam) that suppresses facultative Gram-negative bacteria but not enterococci or anaerobes and does not promote pathogen colonization Urine collected 24 hours after the last treatment was analyzed by LC-MS.
Results: We identified over 25 compounds, many of which had not been previously reported in mouse urine. Eleven small phenolic molecules showed significant antibiotic-related changes. Urinary levels of the hydroxyphenylpropionic acids were suppressed by clindamycin and piperacillin/tazobactam treatment, but were elevated in aztreonam-treated mice. In addition, aztreonam treatment was associated with elevated levels of the dihydroxyhydrocinnamic acids.
Conclusions: Profiles of differential changes in urinary small phenolic molecules may provide an index of anaerobic bacterial species in the GMB and could prove useful in monitoring susceptibility to overgrowth of pathogens such as C. difficile.
Keywords: phenolic acids; microbiota; schizophrenia; autism; anaerobes; mice; clindamycin; aztreonam; piperacillin/tazobactam; antibiotics
Numerous small molecules (< 2,000 Da) produced by the gut microbiota (GMB) [1, 2] enter the systemic circulation and are excreted in urine [3]. Identifying these chemicals and clarifying their underlying kinetics may facilitate development of biomarkers relevant to GMB-sensitive diseases. For example, alteration of the GMB by antibiotics may facilitate colonization by and overgrowth of healthcare-associated pathogens such as Clostridium difficile and vancomycin-resistant enterococci (VRE) [4-6]. C. difficile is the most common pathogen identified as a cause of healthcare-associated infections in the United States, resulting in substantial morbidity, mortality, and economic burden [7]. Having an index for vulnerability or conversely resistance to colonization by pathogens could be clinically useful. To that end, we recently demonstrated that several GMB-associated urinary chemicals were affected by antibiotic treatment in a mouse model of colonization resistance (ie, a host defense provided by the indigenous microbiota to inhibit colonization and overgrowth of potentially pathogenic microorganisms) [8].
The urinary metabolome includes numerous small phenolic molecules (SPMs) that can either be synthesized from L-phenylalanine (L-PHE) or L-tyrosine (L-TYR) [9, 10] or derived from the biotransformation of polyphenols or related dietary constituents [11]. Anaerobic bacteria such as Clostridium species, includingC. difficile, have been implicated in all these routes [11-15]. Furthermore, many of the same SPMs have been identified as potential biomarkers in neuropsychiatric conditions such as schizophrenia and autism [12-15]. Accordingly, our first step was to determine which of the SPMs are affected by targeted perturbations of the GMB.
Liquid chromatography (LC) coupled with mass spectrometry (MS) offers high sensitivity and resolution yet relatively simple sample preparation, and it is now the preferred platform for both targeted and non-targeted metabolomic studies [16]. Even this approach, however, has shortcomings. A prototypical SPM of interest, 3-hydroxy-3-(3-hydroxyphenyl) propanoic acid (3,3-HPHPA),has 12 structural isomers (C9H10O4) differing in the position of a single hydroxyl group [ 17]. The isomers exhibit similar retention times and mass spectra and hence can easily be misidentified by LC/MS [13, 18]. In addition, many SPMs are present in mammalian compartments as mixtures of free chemical and various conjugates [11, 19]. In response, we recently developed an LC-MS protocol that addresses these challenges and supports high resolution and accurate quantification of multiple SPMs [17]. We have now applied this assay to the SPM urinary metabolome of mice with a diverse intestinal microbiota used in a mouse model of colonization resistance [4]. Our primary hypothesis was that urinary SPMs would be differentially affected by antibiotics that do versus do not suppress intestinal anaerobes and promote colonization by healthcare-associated pathogens.
The study protocol was approved by the Cleveland VA Medical Center’s Institutional Animal Care and Use Committee. Female CF-1 mice weighing 25 to 30 g (Harlan Sprague-Dawley, Indianapolis, IN) were housed in groups (n = 5) and fed a sterilized Teklad Global 18% protein-extruded rodent diet (Harlan Teklad, Madison, WI). Groups received daily subcutaneous injections (0.1 mL total volume) of saline, clindamycin (14 mg/day), piperacillin/tazobactam (8 mg/day), or aztreonam (3 mg/day) for 2 consecutive days. The doses were equivalent to clinical doses administered to patients over a 24-hour period (mg/g body weight).
Clindamycin and piperacillin/tazobactam were studied because they have previously been shown to significantly suppress anaerobes in stool and promote colonization by and overgrowth of healthcare-associated pathogens, including C. difficile, VRE, and Klebsiella pneumoniae [5, 6] (Supplementary Table 1). Clindamycin inhibits anaerobes but not indigenous enterococci or facultative gram-negative bacilli (eg, Escherichia coli), whereas piperacillin/tazobactam has broad-spectrum activity resulting in inhibition of indigenous enterococci and facultative gram-negative bacilli in addition to anaerobes [5]. Aztreonam was studied as a comparator because it does not suppress anaerobes in stool or promote pathogen colonization [4, 5]. Aztreonam inhibits indigenous facultative gram-negative bacilli but not enterococci [5].
On day 3, when antibiotic-induced changes in GMB composition were expected to be maximal [6, 8], individual mice were placed in clean cages with no bedding and observed continuously for up to 6 periods of 1 hour. Fresh urine specimens were collected by pipette tip and transferred to sterile Eppendorf tubes. Our preliminary experiments demonstrated that even transient contact of urine with stool could change metabolite concentration in the urine. For that reason, urine specimens in contact with stool pellets were not collected and mice were moved into new clean cages as needed to avoid contamination of urine by stool [8]. Urine specimens were centrifuged at 14,000g for 15 minutes at 8°C, digested with beta-glucuronidase and aryl sulfatase for 24 hours according to the manufacturer’s recommendations (Roche Diagnostics GmbH, Mannheim, Germany) as previously described [17], and then frozen at -80°C until being thawed for analysis [8, 17].
Antibiotics were USP grade (Pfizer, New York). All chemicals and reagents were of the highest purity and grade commercially available. Chemical standards for 3,3-HPHPA and internal standard (2-(Benzoylamino) acetic Acid-d5) (HA-d5) were purchased from Toronto Research Chemicals (North York, Canada), and 3-(2,3-dihydroxyphenyl)propanoic acid (2,3-DHHCA) was procured from Santa Cruz Biotechnology (Santa Cruz, CA). All other standards were purchased from Sigma-Aldrich Co. LLC. (St. Louis, MO). LC-MS grade solvents; water, formic acid, and acetonitrile (ACN) were purchased from Fisher Scientific (Pittsburgh, PA).
A standard stock solution containing all compounds of interest and the internal standards was prepared by accurate weighing of each compound (1 mg/mL) in suitable solvent and initially blanketed with nitrogen in glass vials equipped with Teflon®-lined screw caps and stored at –20°C. The stock solution was serially diluted to obtain working solutions in the range 10 ng/mL to 2000 ng/mL. To generate standard curves over a range of concentrations for quantitation, we used charcoal-stripped urine for the matrix (IRISpec CA/CB reagent, Beckman Coulter, Chatsworth, CA). Both reagents were suitable, and in this case CB reagent was used as the matrix, when exploring matrix effects in urine. Protein was not precipitated, but all cells were eliminated with a clinical spin and prepared the same way as urine samples. In brief, 10 μL of the internal standard at 1000 ng/mL and 50 μL of matrix were added to each diluted standard solution at the final volume of 100 μL (for details see [8]).
Urine samples were stored at -80°C until thawed for analysis. After aliquoting 100 μL of urine, internal standards in water were added, and any protein in samples was precipitated with 6-fold volume of ice-cold acetonitrile by vortexing for 1 minute, and followed by recovery of the supernatant following centrifugation at 14,000 g at 5◦C for 15 minutes. After drying (Speed vacuum), samples were reconstituted to 100 μL of which 20 μL was injected for LC–MS analysis. When digestion was required, supernatants were transferred to a new 1.5-mL Eppendorf tube and digested with aryl sulfatase and beta glucuronidase (from Helix pomatia, Roche Applied Science, Mannheim Germany) per manufacturer instructions [17, 20]. After digestion, the samples were centrifuged (14,000 g × 10 minutes). The supernatant was transferred to a clean HPLC injection vial for LCMS analysis. Sample metabolites were previously evaluated for interfering chemical species known to exert quenching or matrix effects [21]. We previously demonstrated that once the issue of potential conjugates was addressed, urine samples required minimal additional pretreatments (eg, with sulfatase, glucuronidase) [8]. Preliminary stability studies were evaluated using pooled samples in triplicate. Freeze-thaw stability was assessed for 4 cycles from −80◦C to room temperature. Benchtop stability was tested by analyzing samples, which were left out on the bench top at room temperature for more than 8 hours before analysis.
The initial analysis was conducted at the Cleveland Clinic Small Molecule Mass Spectrometry Core on a Shimadzu LCMS-8050 (Kyoto Japan). Compound ions were targeted for collision-induced dissociation (CID) fragmentation based on the previously determined accurate mass and retention time of standards [8, 17]. Analytes were identified by their LC retention time and by monitoring their specific multiple reaction monitoring (MRM) transitions. Selectivity was evaluated by injecting a single analyte, while measuring MRM transitions of all analytes under consideration. These transitions are described in our previous report [17]. Typically, 1 quantitation MRM transition is collected along with several confirmatory ion pairs using a minimum of 2 transitions for each compound, where possible.
A reconstituted sample of 20 μL was injected onto a Shimadzu 8050LCMS system (Shimadzu Kyoto, Japan) with a Luna, C18 (3 μm, ODS 100A, 2 × 150 mm) LC column from (Phenomenex, Rancho Palos Verdes, CA). Eluents A (water + 0.2% acetic acid) and B (acetonitrile + 0.2% acetic acid) were used under gradient conditions at a flow rate of 0.3 mL/min. The gradient program started with isocratic elution with 100% A (0-2 minutes, followed by 100% A to 100% B; 2-8 minutes; 8-18 minutes, 100% B, 18-18.5 minutes, 100% B to 100% A; 18.5-26 minutes, 100% A). This permitted the separation of over 50 analytes. Then 5 μL of HPLC column effluent was introduced onto a Shimadzu 8050 triple quadruple MS system (Shimadzu, Kyoto, Japan) and analyzed using either positive or negative electrospray ionization (ESI) in the MRM mode [17] with the following parameters: nebulizing gas flow 3 L/min, heating gas flow 10 L/minutes, drying gas flow 10 L/minutes, interface temperature 300°C, DL temperature 250°C, heat block temperature 400°C, and argon as the collision-induced dissociation (CID) gas. Retention time was determined during method development and the fragmentation patterns for urine metabolite unknowns and authentic standard compounds were compared after an initial optimization of the collision energy [17]. Internal standard calibration curves were used to quantify metabolites. The use of isotope dilution LC-MS for metabolic profiling is the standard method for quantification of rodent and bacterial metabolites or cellular material [22].
Quantitation was performed using the internal standard calibration method. Each calibration sample was assayed in duplicate over 3 different days. The peak area of each analyte was divided by the peak area of the corresponding internal standard. This ratio was then plotted on the y-axis against known quantities of each analyte to generate calibration curves by linear regression. Whenever possible we used isotope dilution mass spectrometry and standards with a minimum of 3 mass units difference relative to the native compound. Where no isotope was commercially available, we used unlabeled compounds that were structurally similar to the compound of interest. The sensitivity of this method was based on the limits of detection and limits of quantification values for each analyte, which were determined at a signal-to-noise ratio of 3 and 10, respectively. To evaluate the carry-over effect, a blank of methanol and water was injected after the highest concentration standard and after each set of standards and throughout the runs every 20 injections.
Urinary metabolites were normalized using creatinine and the d3-creatinine internal standard as previously described [17]. This required special handling and hydrophilic interaction liquid chromatography (HILIC). HILIC employs a multimodal partitioning technique, where highly polar analytes are retained on the column by partitioning between a water-enriched layer attracted by the polar stationary phase and the solvent, consisting of a mixture of acetonitrile and aqueous buffer in the range of 10% to 90%. Creatinine was quantified using the following parameters: HILIC SeQuant® ZIC®‐cHILIC Column, 250 x 4.6 mm ID 3 μm particle size. Eluent C (acetonitrile) and eluent D (water with 0.1M ammonium acetate) were used across the HILIC gradient (0‐2 minutes, 90% C / 10% D; 2‐20 minutes, 90% C / 10% D to 10% C / 90% D; 20‐21 minutes: 10% C / 90% D to 90% C / 10%; D 21‐30 minutes, 90% C / 10% D) at a flow rate of 1 mL/minute. The pressure maximum was 120 to 200 bar [17].
The data were processed using LabSolutions© (Shimadzu) software. Each peak was manually checked for retention time and proper integration, after applying the auto-quantitation method. Analytes were normalized by using urine creatinine. Levels for each of the 3 antibiotic-treated groups were compared to those for control (saline-treated) using the Mann-Whitney U test. Significance was set at an alpha value of 0.05. No adjustments were made for multiple comparisons.
Urinary creatinine concentrations (569 + 135 nmol/mL) in controls did not differ significantly from that of any other antibiotic-treated group. Accordingly, the concentration of each metabolite was normalized by the individual creatinine value. Table 1 shows the concentrations of the metabolites in the urine of control mice collected 24 hours after completion of 2 consecutive days of subcutaneous injections of saline. Each of the compounds was detected in the urine of control mice. The compounds included 11 that to our knowledge had not been previously identified in mouse urine and 5 compounds that had been previously identified but not quantified in mouse urine.
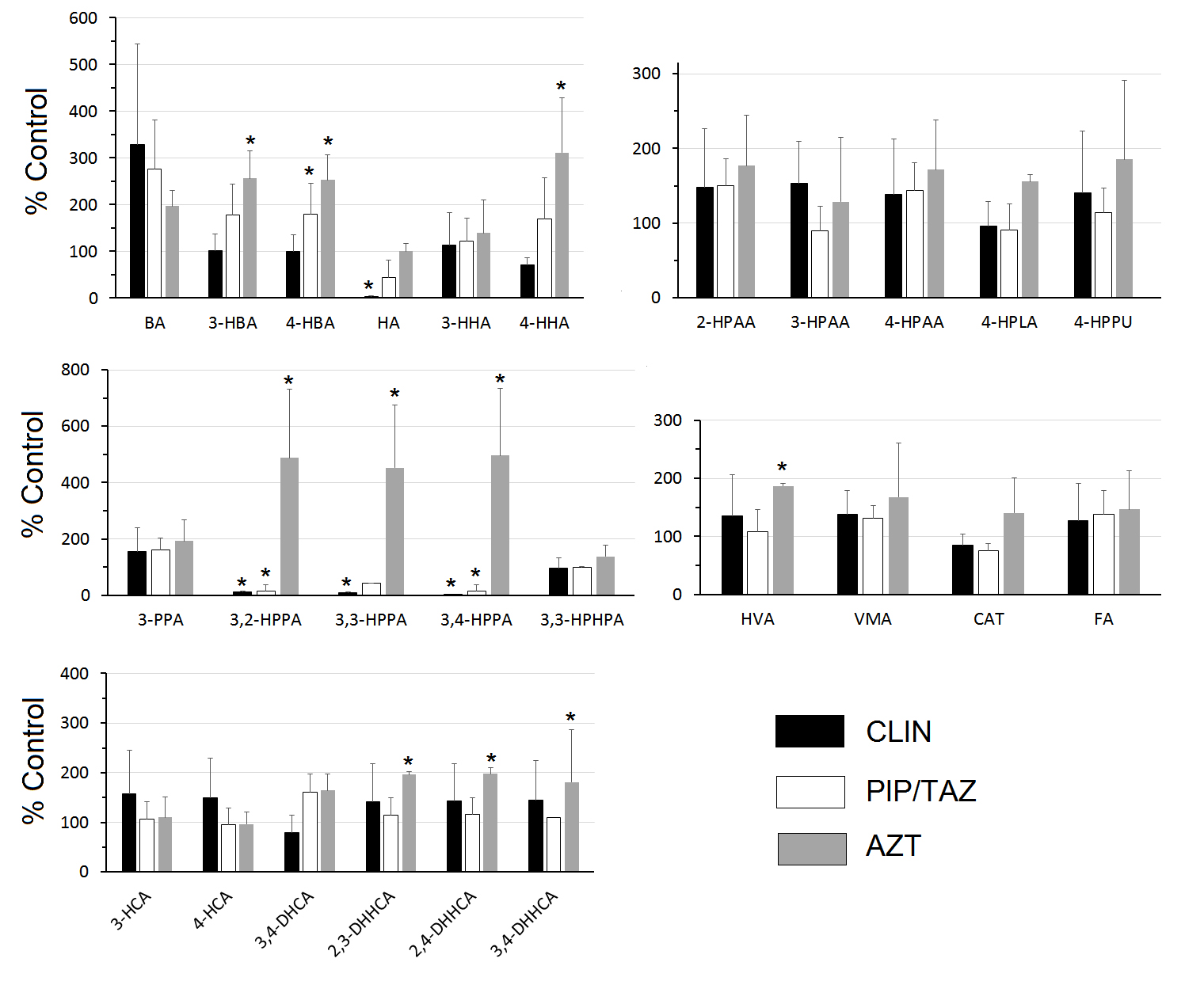
Figure 1 shows the urine concentrations of the SPM metabolites expressed as a percentage of the concentrations in the control mice. Clindamycin treatment resulted in statistically significant reductions in HA and 3,2-HPPA, 3,3-HPPA, and 3,4-HPPA. Piperacillin/tazobactam treatment resulted in statistically significant reductions in 3,2-HPPA and 3,4-HPPA. In contrast, aztreonam treatment resulted in statistically significant increases in 3-hydroxybenzoic acid (3-HBA), 4-HBA, 4-hydroxyhippuric acid, 3,2-HPPA, 3,3-HPPA, 3,4-HPPA, HVA, and the dihydroxyhydrocinnamic acids (DHHCAs) (2,3-DHHCA, 2,4-DHHCA and 3,4-DHHCA) (Figure 1).
|
|
||
|
|
|
|
|
benzoic acid |
0.010 |
0.006 |
|
3-hydroxybenzoic acid† |
14.94 |
1.74 |
|
4-hydroxybenzoic acid |
45.95 |
5.21 |
|
hippuric acid |
433.11 |
139.20 |
|
3-hydroxyhippuric acid† |
1.31 |
0.33 |
|
4-hydroxyhippuric acid |
0.51 |
0.07 |
|
2-hydroxyphenylacetic acid |
12.11 |
2.54 |
|
3-hydroxyphenylacetic acid† |
22.29 |
8.98 |
|
4-hydroxyphenylacetic acid |
11.35 |
2.48 |
|
4-hydroxyphenyllactic acid‡ |
62.82 |
20.52 |
|
4-hydroxyphenylpyruvic acid |
6.44 |
2.02 |
|
3-phenylpropionic acid† |
0.22 |
0.05 |
|
3,2-hydroxyphenylpropionic acid† |
3.37 |
1.69 |
|
3,3-hydroxyphenylpropionic acid‡ |
139.83 |
71.55 |
|
3,4-hydroxyphenylpropionic acid † |
3.35 |
1.87 |
|
3,3-hydroxyphenylhydropropionic acid† |
7.67 |
1.43 |
|
3-hydroxycinnamic acid† |
14.76 |
4.50 |
|
4-hydroxycinnamic acid† |
82.32 |
26.91 |
|
3,4-dihydroxycinnamic acid‡ |
2.08 |
0.87 |
|
2,3-dihydroxyhydrocinnamic acid† |
13.58 |
2.13 |
|
2,4-dihydroxyhydrocinnamic acid† |
14.84 |
2.35 |
|
3,4-dihydroxyhydrocinnamic acid‡ |
6.48 |
1.27 |
|
homovanillic acid |
17.13 |
3.78 |
|
vanillylmandelic acid |
0.24 |
0.04 |
|
ferulic acid‡ |
46.79 |
6.93 |
|
Catechin |
1.94 |
0.67 |
Levels of metabolites in mouse urine collected 24 hours after completion of 2 consecutive days of subcutaneous injections (0.1 mL) of saline. † not previously identified, ‡ previously identified but not quantified in mouse urine under baseline conditions.

Figure 1. Urinary levels expressed as % control. Urine was collected 24 hours after completion of 2 daily SC treatments with saline (control), clindamycin (8 mg/day) (CLIN), piperacillin + tazobactam (8 mg/day) (PIP/TAZO), or aztreonam (3 mg/day) (AZT). Comparisons between each treatment and control were made by Mann-Whitney U test with statistical significance set at P < 0.05* (benzoic acid (BA), 3-hydroxybenzoic acid (3-HBA), 4-hydroxybenzoic acid (4-HBA), hippuric acid (HA), 3-hydroxyhippuric acid (3-HHA), 4-hydroxyhippuric acid (4-HHA), 2-hydroxyphenylacetic acid (2-HPAA), 3-hydroxyphenylacetic acid (3-HPAA), 4-hydroxyphenylacetic acid (4-HPAA), 4-hydroxyphenyllactic acid (4-HPLA), 4-hydroxyphenylpyruvic acid (4-HPPU), 3-phenylpropionic acid (3-PPA), 3,2-hydroxyphenylpropionic acid (3,2-HPPA), 3,3-hydroxyphenylpropionic acid (3,3-HPPA), 3,4-hydroxyphenylpropionic acid (3,4-HPPA 3,3-hydroxyphenylhydropropionic acid (3,3 -HPHPA), 3-hydroxycinnamic acid (3-HCA) 4-hydroxycinnamic acid (4-HCA), 3,4-dihydroxycinnamic acid (3,4-DHCA), 2,3-dihydroxyhydrocinnamic acid (2,3-DHHCA), 2,4-dihydroxyhydrocinnamic acid (2,4-DHHCA), 3,4-dihydroxyhydrocinnamic acid (3,4-DHHCA), homovanillic acid (HVA), vanillylmandelic acid (VMA), ferulic acid (FA), catechin (CAT).
Using an improved LC/MS approach we were able to quantify 25 SPMs and 1 polyphenol (catechin) (Table 1). Eleven of the SPMs were significantly affected by antibiotic treatment. In support of our primary hypothesis, the SPM urinary metabolites were differentially affected by antibiotics depending on their ability to suppress versus not suppress intestinal anaerobes and promote colonization by healthcare-associated pathogens. Specifically, clindamycin and piperacillin/tazobactam are known to suppress anaerobes and treatment with these agents resulted in reductions in the group of hydroxyphenylpropionic acids. In contrast, aztreonam is known to have no or minimal impact on anaerobes, and treatment with this agent did not result in suppression of hydroxyphenylpropionic acids, but it promoted increases in dihydroxyhydrocinnamic acids.
There are several reasons why many of the SPMs have not previously been detected in mouse urine. First, small molecules in biological systems often exist as diverse conjugates generated by the host and/ or gut microbiota. The major phase II conjugation and metabolism pathways include methylation, β-glucuronidation of hydroxyl-, carboxyl-, amino- and thiol-groups, and sulfation of hydroxyl- and amino groups [11, 19]. By conducting hydrolysis using both β-glucuronidase and aryl sulfatase before analyzing samples, we liberated all of a given compound irrespective of its native conjugation status, increasing the net amount of the free chemical and making it easier to measure.
Second, the family of SPMs contains many highly similar molecules. In preliminary studies, the structural isomers of 3,3-HPHPA for instance, could not be adequately resolved using a different column (Prodigy C 18 column (2×150 mm, Prodigy, 5μm, ODS(2), Phenomenex, Rancho Palos Verdes, CA) (data not shown). The 3,3-HPHPA peak was embedded in multiple overlapping isomeric peaks. Through a series of studies, employing new LC column technology (Luna® 3 μm C18(2) 100 Å LC Column 150 x 2 mm), structural and targeted metabolomic approaches across multiple platform systems, and state of the art software in conjunction with MS library searches, we eventually achieved separation and identification of important isomers and fragmentation patterns [17].
Third, the strongest confirmation of molecular identity requires availability of an authentic high purity standard. We were able to purchase commercial standards for all the chemicals we quantified. However, there were several structural isomers (3,2-HPHPA, 3,4-HPHPA, 3-hydroxy-2-(3-hydroxyphenyl)propanoic acid), o -hydroxyphenyllactic acid, m-hydroxyphenyllactic acid, 3-hydroxyphenyllactic acid (2,3-Dihydroxy-3-phenylpropanoic acid), 3,5-DHHCA) that we could not readily obtain and hence, whose presence or absence we could not determine.
The urinary metabolome of the mouse has not been as extensively characterized as that of the rat. However, in agreement with another study in the mouse [23] , HA was by far the most abundant SPM excreted (Table 1). Comparisons with the rat suggest several similarities. Levels of 3,3-HPPA exceed those of 3,4-HPPA by several-fold [24, 25] as do levels of 4-hydroxycinnamic acid relative to 3-hydroxycinnamic acid [26]; There are significant amounts of 4-hydroxyphenyllactic acid [24] (Table 1).
Oral administration of isotopically labeled L-PHE or L-TYR results in excretion of multiple labeled SPMs in man and other mammals [9,27, 28]. The appearance of some of these, can be suppressed by pretreatment with neomycin [9], an antibiotic that is poorly absorbed from the gastrointestinal tract and hence acts only on the GMB [29]. Analogously, orally administered isotopically labeled flavonoids can be converted to numerous labeled urinary SPMs both in man and in the rodent [30-32]. Levels of many SPMs, can be eliminated or significantly lowered by prior administration of neomycin [30, 33] or other broad-spectrum antibiotic treatments [34]. These same SPMs are identified as GMB-related in studies of animals considered to be germ-free [24, 35-38] and in patients with ileostomies [39-41]. Thus, it is now well established that kinetics of some SPMs are at least in part dependent on the GMB [11, 42] (Figure 2), whereas those of other SPMs are largely mediated by endogenous processes within host cells [43].
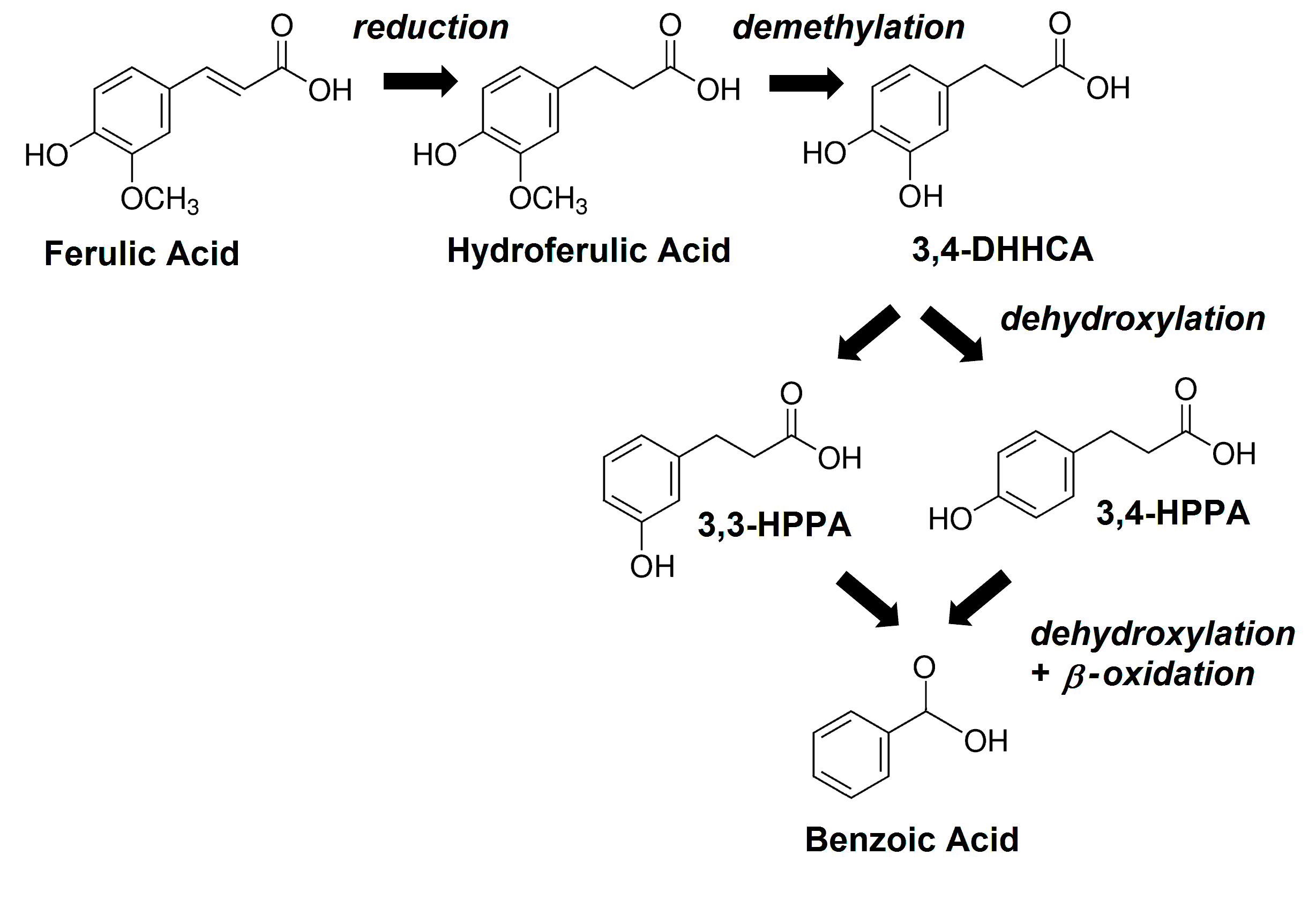
Figure 2. The main GMB-mediated metabolic pathway for ferulic acid in the mouse; benzoic acid (BA), 3,3-hydroxyphenylpropionic acid (3,3-HPPA), 3,4-hydroxyphenylpropionic acid (3,4-HPPA), 3,4-dihydroxyhydrocinnamic acid (3,4-DHHCA).
Diet is another dominant factor that determines the GMB-dependent metabolome [11, 44]. Our mice were fed a largely plant-based laboratory diet consisting of soybean meal, brewer’s dry yeast, and wheat [45]. Soybeans contain only low quantities of catechin and cinnamic acids [46] and brewer’s yeast does not contain polyphenolic precursors of SPMs. Wheat and most other grains do however contain large amounts of ferulic acid (FA) [47, 48]. Less than 10% of the total FA is in the free form that can readily be absorbed in the small intestine [49]. Most FA, like other plant-derived hydrocinnamates, is ester-linked to plant cell wall polymers (cellulose, arabinoxylan, and β-glucan) and cannot readily be absorbed in this complexed form [50, 51]. Esterase activity within the intestinal mucosa or the GMB must release the free FA before it can be absorbed and/or further metabolized [50-52]. In our study, urinary levels of FA were over 20-fold higher than those of catechin (Table 1) and did not vary across antibiotic treatment groups (Figure 1), suggesting that FA was highly bioavailable across treatment groups. In the rat, dietary FA makes a larger contribution to urinary SPAs than does free L-PHE or L-TYR [27, 53].
Different classes of antibiotics produce different metabolomic patterns [8, 54], confirming some selectivity in the contributions that various constituents of the GMB make to the metabolome. An overview of our data provides additional detail. In the clindamycin and piperacillin/tazobactam groups, levels of 3,2-HPPA, 3,3-HPPA, and 3,4-HPPA were markedly lower than in controls (Figure 1). However, those same metabolites were elevated in the aztreonam-treated group, as were 2,3-DHHCA, 2,4-DHHCA, and 3,4-DHHCA. The findings starkly separate the 2 groups of antibiotics. In the rat, the excretion of urinary 3,3-HPPA is dramatically elevated when germ-free rats are inoculated with conventional GMB [24]. Broad spectrum antibiotic treatments lower levels of urinary 3,3-HPPA in mice [55] as well as levels of 3,3-HPPA [34, 55-57] and 3,4-HPPA [56] in rats. This makes the acute increases in 3,2-HPPA, 3,3-HPPA, and 3,4-HPPA (Figure 1) all the more striking. There are insufficient reports to comparably assess the levels of DHHCA. One group reported that a high bran diet, another source of FA, elevated urinary levels of a sulfated-DHHCA isomer, which could not be fully resolved [58]. Our data would indicate that all the DHHCA isomers we measured are differentially sensitive to the GMB (Table 1). The accurate identification of the different DHHCA isomers is important for at least 2 reasons. First, they can be readily mistaken for other structural isomers [13]. Second, 3,4-DHHCA has been identified in rodent brain [59] where it can promote brain plasticity and attenuate some depression-like phenotypes [60].
Hippuric acid is an acyl glycine formed through the conjugation of benzoic acid with glycine [61]. Urinary hippuric acid levels are lower in germ-free mice than conventionally reared controls [1, 36], in mice acutely treated with vancomycin [62], and in rats acutely treated with a variety of antibiotics. This is attributed to GMB influences on benzoic acid synthesis [61]. Thus, the finding that clindamycin dramatically lowered urinary hippuric levels (Figure 1) is hardly surprising. However, in agreement with our earlier report [8], hippuric acid levels in the piperacillin/tazobactam or aztreonam-treated groups were not different from controls (Figure 1), providing additional separation between antibiotics.
This report is the first identification of 3,3-HPHPA in the mouse (Table 1). Of 4 groups that analyzed rat urine for the presence of 3,3-HPHPA, 2 confirmed its presence [24, 63] and 2 did not [32, 38]. One group reported that inoculation of germ-free rats with feces from specific-pathogen-free rats had no effect on urinary 3,3-HPHPA [24]. Similarly, we did not find any effect of antibiotic treatment on 3,3-HPHPA levels in the mouse (Figure 1). The oral antibiotic neomycin has been reported to dramatically lower excretion of 3,3-HPHPA in man [9, 64]. Whether species differences or other factors account for this is unclear.
We evaluated female mice, in keeping with the larger body of work from our laboratory (CJD).
Gender-specific effects of antibiotic on the GMB are certainly known [65]. Effects in male mice would need to be independently determined.
Our data demonstrate that antibiotic treatments that target different groups of bacteria, produce significantly different urinary phenolic metabolomes (Figure 1). We previously showed that a course of piperacillin/tazobactam but not aztreonam disrupted colonization resistance to C. difficile [8]. Clindamycin is another antibiotic treatment that disrupts colonization resistance in mice [6]. The similarities in SPM changes in the clindamycin and piperacillin/tazobactam groups as opposed to the aztreonam-treated group and controls, raise the possibility that an elevation of urinary 3,2-HPPA, 3,3-HPPA, and 3,4-HPPA may reliably track antibiotic-induced colonization resistance. Such changes in the phenolic metabolome, combined with previously reported findings [8] may support the development of a clinical panel that could distinguish an intact flora from one vulnerable to overgrowth by pathogenic anaerobic bacteria. In support of this, longitudinal studies should be conducted that concurrently characterize the urinary phenolic metabolome, the fecal metabolome, the fecal microbiota, and functional measures of colonization resistance during the period of antibiotic treatment and recovery.
The first author performed more than 90% of the work, which was supported by the Louis Stokes Cleveland DVAMC Research Service and by a grant from the VISN 10 Research Initiative Program (to CJD and GEJ).
The authors thank The Parker Hannifin Corporation for refurbishing our nitrogen generator, Paul Minkler for helpful discussion and technical assistance, and Engine Industries for assistance with Figures and visual formatting requirements.
The manuscript content is solely the responsibility of the authors and does not necessarily represent the official views of the Department of Veterans Affairs, the authors’ employers, or their academically affiliated institutions.
Curtis J. Donskey has received research funding from Clorox, GOJO, Pfizer, Avery Dennison, PDI, and Boehringer Laboratories. He is also an associate editor for Pathogens and Immunity. All other authors report no relevant conflicts of interest.
1. Wikoff WR, Anfora AT, Liu J, Schultz PG, Lesley SA, Peters EC, Siuzdak G. Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(10):3698-703. PubMed PMID: 19234110. Pubmed Central PMCID: PMC2656143. doi: 10.1073/pnas.0812874106
2. Collins SM, Surette M, Bercik P. The interplay between the intestinal microbiota and the brain. Nat Rev Microbiol. 2012;10(11):735-42.
3. Wishart DS, Feunang YD, Marcu A, Guo AC, Liang K, Vazquez-Fresno R, Sajed T, Johnson D, Li C, Karu N, Sayeeda Z, Lo E, Assempour N, Berjanskii M, Singhal S, Arndt D, Liang Y, Badran H, Grant J, Serra-Cayuela A, Liu Y, Mandal R, Neveu V, Pon A, Knox C, Wilson M, Manach C, Scalbert A. HMDB 4.0: the human metabolome database for 2018. Nucleic Acids Research. 2018;46(D1):D608-d17. PubMed PMID: 29140435. Pubmed Central PMCID: PMC5753273. doi: 10.1093/nar/gkx1089
4. Pultz NJ, Donskey CJ. Effect of antibiotic treatment on growth of and toxin production by Clostridium difficile in the cecal contents of mice. Antimicrobial Agents and Chemotherapy. 2005;49(8):3529-32. PubMed PMID: 16048976. Pubmed Central PMCID: PMC1196291. doi: 10.1128/aac.49.8.3529-3532.2005
5. Pultz NJ, Stiefel U, Subramanyan S, Helfand MS, Donskey CJ. Mechanisms by which anaerobic microbiota inhibit the establishment in mice of intestinal colonization by vancomycin-resistant Enterococcus. J Infect Dis. 2005;191(6):949-56.
6. Jump RL, Polinkovsky A, Hurless K, Sitzlar B, Eckart K, Tomas M, Deshpande A, Nerandzic MM, Donskey CJ. Metabolomics analysis identifies intestinal microbiota-derived biomarkers of colonization resistance in clindamycin-treated mice. PloS One. 2014;9(7):e101267. PubMed PMID: 24988418. Pubmed Central PMCID: PMC4079339. doi: 10.1371/journal.pone.0101267
7. Deshpande A, Pant C, Olyaee M, Donskey CJ. Hospital readmissions related to Clostridium difficile infection in the United States. American Journal of Infection Control. 2018;46(3):346-7. PubMed PMID: 29050906. doi: 10.1016/j.ajic.2017.08.043
8. Obrenovich ME, Tima M, Polinkovsky A, Zhang R, Emancipator SN, Donskey CJ. Targeted Metabolomics Analysis Identifies Intestinal Microbiota-Derived Urinary Biomarkers of Colonization Resistance in Antibiotic-Treated Mice. Antimicrobial Agents and Chemotherapy. 2017;61(8):pii: e00477-17. PubMed PMID: 28584146. Pubmed Central PMCID: PMC5527637. doi: 10.1128/aac.00477-17
9. Curtius HC, Redweik U, Steinmann B, Leimbacher W, Wegmann H. Use of deuterated tyrosine and phenylalanine in the study of catecholamine and aromatic amino acid metabolism. In: Klein ER, Klein PD, editors. Proceedings of the Second International Conference on Stable Isotopes, October 20-23, 1975, Oak Brook, Illinois. Washington, DC: U.S. Energy Research and Development Administration; 1976. p. 385-91.
10. Rampini S, Vollmin JA, Bosshard HR, Muller M, Curtius HC. Aromatic acids in urine of healthy infants, persistent hyperphenylalaninemia, and phenylketonuria, before and after phenylalanine load. Pediatr Res. 1974;8(7):704-9.
11. Williamson G, Clifford MN. Role of the small intestine, colon, and microbiota in determining the metabolic fate of polyphenols. Biochemical Pharmacology. 2017;139:24-39. PubMed PMID: 28322745. doi: 10.1016/j.bcp.2017.03.012
12. Kesli R, Gokcen C, Bulug U, Terzi Y. Investigation of the relation between anaerobic bacteria genus clostridium and late-onset autism etiology in children. J Immunoassay Immunochem. 2014;35(1):101-9.
13. Shaw W. Increased urinary excretion of a 3-(3-hydroxyphenyl)-3-hydroxypropionic acid (HPHPA), an abnormal phenylalanine metabolite of Clostridia spp. in the gastrointestinal tract, in urine samples from patients with autism and schizophrenia. Nutritional Neuroscience. 2010;13(3):135-43. PubMed PMID: 20423563. doi: 10.1179/147683010X12611460763968
14. Shaw W. Clostridia bacteria in the gastrointestinal tract as a major cause of depression and other neuropsychiatric disorders. In: Greenblatt J, Brogan K, editors. Integrative Psychiatry for Depression: Redefining Models for Assessment, Treatment, and Prevention of Mood Disorders. New York, NY: Taylor and Francis; 2016. p. 31-48.
15. Xiong X, Liu D, Wang Y, Zeng T, Peng Y. Urinary 3-(3-Hydroxyphenyl)-3-hydroxypropionic Acid, 3-Hydroxyphenylacetic Acid, and 3-Hydroxyhippuric Acid Are Elevated in Children with Autism Spectrum Disorders. Biomed Res Int. 2016;2016:9485412. PubMed PMID: 27123458. Pubmed Central PMCID: PMC4829699. doi: 10.1155/2016/9485412
16. Fang ZZ, Gonzalez FJ. LC-MS-based metabolomics: an update. Archives of Toxicology. 2014;88(8):1491-502. PubMed PMID: 24710571. doi: 10.1007/s00204-014-1234-6
17. Obrenovich ME, Donskey CJ, Schiefer IT, Bongiovanni R, Li L, Jaskiw GE. Quantification of phenolic acid metabolites in humans by LC-MS: a structural and targeted metabolomics approach. Bioanalysis. 2018;10(19):1591-608. PubMed PMID: 30295550. doi: 10.4155/bio-2018-0140
18. Bouatra S, Aziat F, Mandal R, Guo AC, Wilson MR, Knox C, Bjorndahl TC, Krishnamurthy R, Saleem F, Liu P, Dame ZT, Poelzer J, Huynh J, Yallou FS, Psychogios N, Dong E, Bogumil R, Roehring C, Wishart DS. The human urine metabolome. PloS One. 2013;8(9):e73076. PubMed PMID: 24023812. Pubmed Central PMCID: PMC3762851. doi: 10.1371/journal.pone.0073076
19. Shangari N, Chan TS, O'Brien PJ. Sulfation and glucuronidation of phenols: implications in coenyzme Q metabolism. Methods in Enzymology. 2005;400:342-59. PubMed PMID: 16399359. doi: 10.1016/s0076-6879(05)00020-0
20. Obrenovich ME, Donskey CJ, Schiefer IT, Bongiovanni R, Li L, Jaskiw GE. Quantification of Phenolic Acid Metabolites in Humans by LC-MS – A Structural and Targeted Metabolomics Approach. Bioanalysis.(in press).
21. van de Merbel NC. Quantitative determination of endogenous compounds in biological samples using chromatographic techniques. Trends Anal Chem. 2008;27(10):924-33.
22. Wu Y, Li L. Development of isotope labeling liquid chromatography-mass spectrometry for metabolic profiling of bacterial cells and its application for bacterial differentiation. Analytical Chemistry. 2013;85(12):5755-63. PubMed PMID: 23495969. doi: 10.1021/ac400330z
23. Vostalova J, Galandakova A, Palikova I, Ulrichova J, Dolezal D, Lichnovska R, Vrbkova J, Rajnochova Svobodova A. Lonicera caerulea fruits reduce UVA-induced damage in hairless mice. Journal of Photochemistry and Photobiology B: Biology. 2013;128:1-11. PubMed PMID: 23974431. doi: 10.1016/j.jphotobiol.2013.07.024
24. Goodwin BL, Ruthven CR, Sandler M. Gut flora and the origin of some urinary aromatic phenolic compounds. Biochemical Pharmacology. 1994;47(12):2294-7. PubMed PMID: 8031324.
25. Gasperotti M, Masuero D, Guella G, Mattivi F, Vrhovsek U. Development of a targeted method for twenty-three metabolites related to polyphenol gut microbial metabolism in biological samples, using SPE and UHPLC-ESI-MS/MS. Talanta. 2014;128:221-30. PubMed PMID: 25059152. doi: 10.1016/j.talanta.2014.04.058
26. Khanal R, Howard LR, Prior RL. Urinary excretion of phenolic acids in rats fed cranberry, blueberry, or black raspberry powder. Journal of Agricultural and Food Chemistry. 2014;62(18):3987-96. PubMed PMID: 24180593. doi: 10.1021/jf403883r
27. Armstrong MD, Chao FC, Parker VJ, Wall PE. Endogenous formation of hippuric acid. Proceedings of the Society for Experimental Biology and Medicine. 1955;90(3):675-9. PubMed PMID: 13289873.
28. Bernhard K, Vuilleumier JP, Brubacher G. Zur Frage der Entstehung der Benzoesäure im Tierkörper. Helvetica Chimica Acta. 1955;38(6):1438-44. doi: 10.1002/hlca.19550380616
29. Breen KJ, Bryant RE, Levinson JD, Schenker S. Neomycin absorption in man. Studies of oral and enema administration and effect of intestinal ulceration. Annals of Internal Medicine. 1972;76(2):211-8. PubMed PMID: 5009591.
30. Baba S, Furuta T, Fujioka M, Goromaru T. Studies on drug metabolism by use of isotopes XXVII: urinary metabolites of rutin in rats and the role of intestinal microflora in the metabolism of rutin. J Pharm Sci. 1983;72(10):1155-8.
31. Ottaviani JI, Borges G, Momma TY, Spencer JP, Keen CL, Crozier A, Schroeter H. The metabolome of [2-(14)C](-)-epicatechin in humans: implications for the assessment of efficacy, safety, and mechanisms of action of polyphenolic bioactives. Scientific Reports. 2016;6:29034. PubMed PMID: 27363516. Pubmed Central PMCID: PMC4929566. doi: 10.1038/srep29034
32. Borges G, van der Hooft JJJ, Crozier A. A comprehensive evaluation of the [2-14C](-)-epicatechin metabolome in rats. Free Radical Biology and Medicine. 2016;99:128-38. PubMed PMID: 27495388. doi: 10.1016/j.freeradbiomed.2016.08.001
33. Nakagawa Y, Shetlar MR, Wender SH. Urinary products from quercetin in neomycin-treated rats. Biochimica et Biophysica Acta. 1965;97:233-41. PubMed PMID: 14292832.
34. Griffiths LA. Studies on flavonoid metabolism. Identification of the metabolities of (+)-catechin in rat urine. Biochemical Journal. 1964;92(1):173-9. PubMed PMID: 5840958. Pubmed Central PMCID: PMC1215455.
35. Hanske L, Loh G, Sczesny S, Blaut M, Braune A. The bioavailability of apigenin-7-glucoside is influenced by human intestinal microbiota in rats. Journal of Nutrition. 2009;139(6):1095-102. PubMed PMID: 19403720. doi: 10.3945/jn.108.102814
36. Claus SP, Tsang TM, Wang Y, Cloarec O, Skordi E, Martin FP, Rezzi S, Ross A, Kochhar S, Holmes E, Nicholson JK. Systemic multicompartmental effects of the gut microbiome on mouse metabolic phenotypes. Molecular Systems Biology. 2008;4:219. PubMed PMID: 18854818. Pubmed Central PMCID: PMC2583082. doi: 10.1038/msb.2008.56
37. Goldin BR, Peppercorn MA, Goldman P. Contributions of host and intestinal microflora in the metabolism of L-dopa by the rat. Journal of Pharmacology and Experimental Therapeutics. 1973;186(1):160-6. PubMed PMID: 4723308.
38. Borud O, Midtvedt T, Gjessing LR. Urinary phenolic compounds in gnotobiotic and conventional rats on a free diet, and before and after L-DOPA loading on a milk diet. Acta Pharmacologica et Toxicologica. 1971;30(3):185-92. PubMed PMID: 5003294.
39. Stalmach A, Edwards CA, Wightman JD, Crozier A. Colonic catabolism of dietary phenolic and polyphenolic compounds from Concord grape juice. Food & Function. 2013;4(1):52-62. PubMed PMID: 22961385. doi: 10.1039/c2fo30151b
40. Clifford MN, Jaganath IB, Ludwig IA, Crozier A. Chlorogenic acids and the acyl-quinic acids: discovery, biosynthesis, bioavailability and bioactivity. Natural Product Reports. 2017;34(12):1391-421. PubMed PMID: 29160894. doi: 10.1039/c7np00030h
41. Olthof MR, Hollman PC, Katan MB. Chlorogenic acid and caffeic acid are absorbed in humans. Journal of Nutrition. 2001;131(1):66-71. PubMed PMID: 11208940.
42. Pekkinen J, Rosa NN, Savolainen OI, Keski-Rahkonen P, Mykkanen H, Poutanen K, Micard V, Hanhineva K. Disintegration of wheat aleurone structure has an impact on the bioavailability of phenolic compounds and other phytochemicals as evidenced by altered urinary metabolite profile of diet-induced obese mice. Nutrition & Metabolism. 2014;11(1):1. PubMed PMID: 24383425. Pubmed Central PMCID: PMC3891979. doi: 10.1186/1743-7075-11-1
43. Kanehisa M, Furumichi M, Tanabe M, Sato Y, Morishima K. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Research. 2017;45(D1):D353-d61. PubMed PMID: 27899662. Pubmed Central PMCID: PMC5210567. doi: 10.1093/nar/gkw1092
44. Phipps AN, Stewart J, Wright B, Wilson ID. Effect of diet on the urinary excretion of hippuric acid and other dietary-derived aromatics in rat. A complex interaction between diet, gut microflora and substrate specificity. Xenobiotica. 1998;28(5):527-37. PubMed PMID: 9622854. doi: 10.1080/004982598239443
45. Envigo. Teklad Global 18% Protein Extruded Rodent Diet (Sterilizable) 2018 [cited 2018 08/08/2018]. Diet composition]. Available from: https://www.envigo.com/resources/data-sheets/2018sx-datasheet-0915.pdf .
46. Xu B, Chang SK. Characterization of phenolic substances and antioxidant properties of food soybeans grown in the North Dakota-Minnesota region. Journal of Agricultural and Food Chemistry. 2008;56(19):9102-13. PubMed PMID: 18781761. doi: 10.1021/jf801451k
47. Zhao Z, Egashira Y, Sanada H. Phenolic antioxidants richly contained in corn bran are slightly bioavailable in rats. Journal of Agricultural and Food Chemistry. 2005;53(12):5030-5. PubMed PMID: 15941352. doi: 10.1021/jf050111n
48. Mattila P, Pihlava JM, Hellstrom J. Contents of phenolic acids, alkyl- and alkenylresorcinols, and avenanthramides in commercial grain products. Journal of Agricultural and Food Chemistry. 2005;53(21):8290-5. PubMed PMID: 16218677. doi: 10.1021/jf051437z
49. Mateo Anson N, Aura AM, Selinheimo E, Mattila I, Poutanen K, van den Berg R, Havenaar R, Bast A, Haenen GR. Bioprocessing of wheat bran in whole wheat bread increases the bioavailability of phenolic acids in men and exerts antiinflammatory effects ex vivo. Journal of Nutrition. 2011;141(1):137-43. PubMed PMID: 21106920. doi: 10.3945/jn.110.127720
50. Aura AM. Microbial metabolism of dietary phenolic compounds in the colon. Phytochemistry Reviews. 2008;7(3):407-29.
51. Bondia-Pons I, Aura A-M, Vuorela S, Kolehmainen M, Mykkanen H, Poutanen K. Rye phenolics in nutrition and health. Journal of Cereal Science. 2009;49:323–36. doi: 10.1016/j.jcs.2009.01.007
52. Andreasen MF, Kroon PA, Williamson G, Garcia-Conesa MT. Esterase activity able to hydrolyze dietary antioxidant hydroxycinnamates is distributed along the intestine of mammals. Journal of Agricultural and Food Chemistry. 2001;49(11):5679-84. PubMed PMID: 11714377.
53. Teuchy H, Van Sumere CF. The metabolism of (1- 14 C) phenylalanine, (3- 14 C) cinnamic acid and (2- 14 C) ferulic acid in the rat. Archives Internationales de Physiologie et de Biochimie. 1971;79(3):589-618. PubMed PMID: 4107877.
54. Behr C, Kamp H, Fabian E, Krennrich G, Mellert W, Peter E, Strauss V, Walk T, Rietjens I, van Ravenzwaay B. Gut microbiome-related metabolic changes in plasma of antibiotic-treated rats. Archives of Toxicology. 2017;91(10):3439-54. PubMed PMID: 28337503. doi: 10.1007/s00204-017-1949-2
55. Zheng X, Xie G, Zhao A, Zhao L, Yao C, Chiu NH, Zhou Z, Bao Y, Jia W, Nicholson JK, Jia W. The footprints of gut microbial-mammalian co-metabolism. Journal of Proteome Research. 2011;10(12):5512-22. PubMed PMID: 21970572. doi: 10.1021/pr2007945
56. Swann JR, Tuohy KM, Lindfors P, Brown DT, Gibson GR, Wilson ID, Sidaway J, Nicholson JK, Holmes E. Variation in antibiotic-induced microbial recolonization impacts on the host metabolic phenotypes of rats. Journal of Proteome Research. 2011;10(8):3590-603. PubMed PMID: 21591676. doi: 10.1021/pr200243t
57. Lee SH, An JH, Park HM, Jung BH. Investigation of endogenous metabolic changes in the urine of pseudo germ-free rats using a metabolomic approach. Journal of Chromatography B: Analytical Technologies in the Biomedical and Life Sciences. 2012;887-888:8-18. PubMed PMID: 22300547. doi: 10.1016/j.jchromb.2011.12.030
58. Pekkinen J, Rosa-Sibakov N, Micard V, Keski-Rahkonen P, Lehtonen M, Poutanen K, Mykkanen H, Hanhineva K. Amino acid-derived betaines dominate as urinary markers for rye bran intake in mice fed high-fat diet--A nontargeted metabolomics study. Molecular Nutrition & Food Research. 2015;59(8):1550-62. PubMed PMID: 25944556. doi: 10.1002/mnfr.201500066
59. Gu L, House SE, Rooney L, Prior RL. Sorghum bran in the diet dose dependently increased the excretion of catechins and microbial-derived phenolic acids in female rats. Journal of Agricultural and Food Chemistry. 2007;55(13):5326-34. PubMed PMID: 17536823. 10.1021/jf070100p
60. Wang J, Hodes GE, Zhang H, Zhang S, Zhao W, Golden SA, Bi W, Menard C, Kana V, Leboeuf M, Xie M, Bregman D, Pfau ML, Flanigan ME, Esteban-Fernandez A, Yemul S, Sharma A, Ho L, Dixon R, Merad M, Han MH, Russo SJ, Pasinetti GM. Epigenetic modulation of inflammation and synaptic plasticity promotes resilience against stress in mice. Nat Commun. 2018;9(1):477. PubMed PMID: 29396460. Pubmed Central PMCID: PMC5797143. doi: 10.1038/s41467-017-02794-5
61. Lees HJ, Swann JR, Wilson ID, Nicholson JK, Holmes E. Hippurate: the natural history of a mammalian-microbial cometabolite. Journal of Proteome Research. 2013;12(4):1527-46. PubMed PMID: 23342949. doi: 10.1021/pr300900b
62. Yap IK, Li JV, Saric J, Martin FP, Davies H, Wang Y, Wilson ID, Nicholson JK, Utzinger J, Marchesi JR, Holmes E. Metabonomic and microbiological analysis of the dynamic effect of vancomycin-induced gut microbiota modification in the mouse. Journal of Proteome Research. 2008;7(9):3718-28. PubMed PMID: 18698804. doi: 10.1021/pr700864x
63. Liu H, Tayyari F, Edison AS, Su Z, Gu L. NMR-based metabolomics reveals urinary metabolome modifications in female Sprague-Dawley rats by cranberry procyanidins. The Journal of Nutritional Biochemistry. 2016;34:136-45. PubMed PMID: 27309592. doi: 10.1016/j.jnutbio.2016.05.007
64. Shaw KNF, Gutenstein M, Jepson JB, editors. Intestinal flora and diet in relation to m-hydroxyphenyl acids of human urine. Fifth International Congress of Biochemistry; 1961; Moscow. New York: Pergamon Press; 1963.
65. Harrison CA, Laubitz D, Midura-Kiela MT, Jamwal DR, Besselsen DG, Ghishan FK, Kiela PR. Sexual Dimorphism in the Response to Broad-spectrum Antibiotics During T Cell-mediated Colitis. Journal of Crohn's & Colitis. 2019;13(1):115-26. PubMed PMID: 30252029. Pubmed Central PMCID: PMC6302957. doi: 10.1093/ecco-jcc/jjy144
|
|
|
|
|
|
Type |
lincosamide |
ureidopenicillin / β-lactam/β-lactamase inhibitor |
monobactam |
|
Suppression of indigenous microbiota |
gram (-) and gram (+) anaerobes |
gram (-) and gram (+) anaerobes, enterococci, and gram (-) facultative anaerobes (eg E. coli) |
gram (-) facultative anaerobes (eg E. coli) |
|
Promotion of pathogens |
C. difficile , vancomycin-resistant enterococci, and facultative gram (-) bacilli (e.g., Klebsiella pneumoniae) |
C. difficile , vancomycin-resistant enterococci, and facultative gram (-) negative bacilli (eg, Klebsiella pneumoniae) |
- |