Dominic Paquin-Proulx1, Fabio E. Leal1, Cassia G. Terrassani Silveira2, Alvino Maestri2, Claudia Brockmeyer1, Shannon M. Kitchen1, Vinicius D. Cabido1, Esper G. Kallas2#, Douglas F. Nixon1#
# Co-senior authors
Douglas F. Nixon
Department of Microbiology
Immunology & Tropical Medicine
The George Washington University
20037 Washington, D.C.
202-994-3532
dnixon@gwu.edu
Background: The outbreak of Zika virus (ZIKV) infection in Brazil has raised concerns that infection during pregnancy could cause microcephaly and other severe neurodevelopmental malformations in the fetus. The mechanisms by which ZIKV causes fetal abnormalities are largely unknown. The importance of pre-infection with dengue virus (DENV), or other flaviviruses endemic to Brazil, remains to be investigated. It has been reported that antibodies directed against DENV can increase ZIKV infectivity by antibody dependent enhancement (ADE), suggesting that a history of prior DENV infection might worsen the outcome of ZIKV infection.
Methods: We used bioinformatics tools to design 18 peptides from the ZIKV envelope containing predicted HLA-I T-cell epitopes and investigated T-cell cross-reactivity between ZIKV-infected individuals and DENV-vaccinated subjects by IFNg ELISPOT.
Results: Three peptides induced IFNg production in both ZIKV-infected subjects and in DENV-vaccinated individuals. Flow cytometry indicated that 1 ZIKV peptide induced a CD4+ T-cell response in DENV-vaccinated subjects.
Conclusions: We demonstrated that vaccination against DENV induced a T-cell response against ZIKV and identified one such CD4+ T-cell epitope. The ZIKV-reactive CD4+ T cells induced by DENV vaccination and identified in this study could contribute to the appearance of cross-reactive antibodies mediating ADE.
Keywords: zika virus, dengue virus, T cell, IFNg, ELISPOT, CD4, antibody dependent enhancement
ZIKV: Zika virus
YF: Yellow Fever
DENV: Dengue virus
ADE: Antibody dependent enhancement
OAS: Original antigenic sin
nd: Not done
na: Not applicable
unk: Unknown
Zika virus (ZIKV) infection was first identified in the Ziika forest of Uganda in 1947 [1]. This virus is a member of the Flaviviridae, a family of viruses that also includes Yellow Fever (YF), West Nile, and Dengue viruses (DENV), and ZIKV can be transmitted via mosquito bites, through sexual contact or blood transfusions, or from an infected pregnant woman to her developing fetus. Until recently, ZIKV was understudied because the infection was thought to be associated only with a mild viral illness of limited duration. It is estimated that only 20% of infected humans show signs of ZIKV infection during the acute phase, with symptoms including skin rash, headache, asthenia, and conjunctivitis. In 2014, the virus suddenly expanded its range dramatically and appeared in the Americas, leading to the most widespread ZIKV outbreak recorded. The recent epidemic of ZIKV in Brazil has raised concerns that ZIKV infection during pregnancy could cause severe neurodevelopmental malformations in the fetus, including microcephaly [2, 3]. Evidence includes epidemiological studies showing a strong temporal and geographical association between ZIKV and microcephaly[4], detection of ZIKV in the brain of infants born with microcephaly [3, 5 ,6], and ZIKV neurological damage in animal models [7-9]. Three different studies performed in French Polynesia (retrospective), Rio de Janeiro, and Bahia have estimated the risk of microcephaly to be between 0.95% and 29% for pregnant women infected with ZIKV [10-13]. A more recent study in the United States estimated the risk to be 6%, with an increased vulnerability if ZIKV infection occurred during the first trimester [14]. A temporal association between an increase in the incidence of microcephaly cases and ZIKV infection was also noted in Colombia [15]. The mechanisms by which ZIKV causes fetal abnormalities are largely unknown. In fact, we still do not know the risk factors associated with fetal microcephaly among pregnant women.
The importance of pre-infection with DENV, another flavivirus endemic to Brazil, remains to be investigated. It has been reported that antibodies directed against DENV can increase ZIKV infectivity in vitro by antibody dependent enhancement (ADE) [16-19], suggesting that a history of prior DENV infection might worsen the outcome of a subsequent ZIKV infection. However, the clinical relevance of ADE to ZIKV pathology remains to be determined [20]. Several studies have suggested the possible involvement of cross-reactive T-cell epitopes in DENV pathogenesis. More specifically, memory T-cell clones generated during primary dengue infection in response to epitopes from 1 dengue serotype can cross-react with epitope variants presented during a subsequent infection with a different dengue serotype. We have previously shown differential serotype-specific CD8+ T-cell immunogenicity of DENV proteins [21]. In mice previously infected with DENV, the CD8 T-cell response to ZIKV infection was almost exclusively driven by a limited number of ZIKV/DENV cross-reactive memory cells [22]. To date, no ZIKV or ZIKV/DENV cross-reactive epitopes have been identified in humans. Here, we hypothesized that prior DENV vaccination impacts immunity to ZIKV through T-cell cross-reactivity.
In this study, we investigated virus specific T-cell cross-reactivity between ZIKV and DENV. We selected 18 peptides from the ZIKV envelope with predicted epitopes for the most common HLA-I alleles in Brazil, based on their sequence similarity/dissimilarity with DENV. We identified 3 peptides that induced an IFNg response in ZIKV-infected patients and in DENV-vaccinated individuals. One of these peptides was found to be a CD4+ T-cell epitope.
We selected 7 ZIKV-infected and 9 DENV-vaccinated individuals from São Paulo, Brazil. All participants were enrolled after signing a written consent form approved by the University of São Paulo’s Institutional Review Board (CAPPesq 0652/09 and 553/08, respectively). Samples from the DENV-vaccinated individuals were collected between January and August 2014 before the ZIKV outbreak reached São Paulo and selected based on the expression of at least 1 allele of the following HLA-I: HLA-A2, HLA-A24, and HLA-B44. Of the ZIKV-infected patients, 2 reported previous DENV infection (801 and 533). None of the DENV-vaccinated individuals reported previous DENV infection. Blood from control subjects was obtained from the New York Blood Bank. Peripheral blood mononuclear cells (PBMCs) were isolated by density-gradient sedimentation using Ficoll-Paque (Lymphoprep, Nycomed Pharma, Oslo, Norway). Isolated PBMCs were washed twice in Hank’s balanced salt solution (Gibco, Grand Island, NY) and cryopreserved in RPMI 1640 (Gibco), supplemented with 20% heat-inactivated fetal bovine serum (FBS; Hyclone Laboratories, Logan UT), 50 U/ml of penicillin (Gibco), 50 mg/ml of streptomycin (Gibco), 10mM glutamine (Gibco) and 10% dimethylsulfoxide (DMSO; Sigma, St Louis, MO). Cryopreserved cells from all participants were stored in liquid nitrogen until used in the assays.
DNA was extracted from peripheral blood using Qiagen kits (QIAamp DNA Mini Kit, Qiagen, Inc, Valencia, CA). The HLA A, B, C and DRB1 typing was performed using PCR and amplification using Sequence-Specific Oligonucleotide (SSP) contained in LABType kits (One Lambda, Inc, Canoga Park, CA).
The HLA-I epitope predictions were made using Net CTL 1.2 [23] (with a prediction score threshold for epitope identification of 0.75), and HLA-II binding predictions were made using the IEDB analysis resource consensus tool [24, 25]. Amino acid sequences were aligned using the EMBL-EBI bioinformatics framework [26, 27]. Sequence data from Zika MR766 (NCBI Reference Sequence: YP_002790881.1), DENV-1 (UniProtKB/Swiss-Prot: Q1L6I3), DENV-2 (UniProtKB/Swiss-Prot: B8QED9), DENV-3 (UniProtKB/Swiss-Prot: Q2YH64), and DENV-4 (UniProtKB/Swiss-Prot: Q5UU53) were used.
Cryopreserved PBMC specimens were thawed, washed, and the cell viability was assessed using the Countess Automated Cell Counter system (Invitrogen, Carlsbad, CA). IFNg ELISPOT assays were conducted following the manufacturer’s instructions (Mabtech, Cincinnati, OH). The ZIKV peptides (GenScript, Piscataway, NJ) were reconstituted with DMSO or water at a concentration of 1 mg/ml and used at a final concentration of 10 μg/mL in duplicate. The PBMCs were plated at a concentration of 1 × 105 cells per well. Spots were counted using AID EliSpot Reader software v6.0. Results from unstimulated PBMCs were used to determine the background level of IFNg production and multiplied by 2 for each donor individually.
Freshly thawed PBMCs were labeled with CellTrace Violet (Invitrogen) following the manufacturer’s instructions and cultured for 6 days at 37°C and 5% CO2 in RPMI medium supplemented with 10% fetal bovine serum (FBS) and 20 units/ml of IL-2 (Sigma), in the presence or absence of 10 μg/ml of peptide. The PBMCs were then re-stimulated with peptide in the presence of monensin (Golgi Stop, BD Biosciences, San Jose, CA) for 12 hours. Cells were washed and stained in Brilliant Violet Stain Buffer (BD Biosciences) at room temperature for 15 minutes in 96-well V-bottom plates in the dark. Samples were then washed and fixed using Cytofix/Cytoperm (BD Biosciences) before flow cytometry analysis. Intracellular staining was performed in Perm/Wash (BD Biosciences), and the following mAbs were used, CD3 PerCP-Cy5.5 (clone UCHT1), CD4 BV605 (clone RPA-T4), CD8 BV711 (clone RPA-T8), and IFNg APC (clone B27), all from BD Biosciences. Live/dead Fixable Violet Cell Stain was from Life Technologies (Eugene, OR). All data was acquired on a BD LSRFortessa X-20 instrument (BD Biosciences) and analyzed using FlowJo Version 9.9.4 software (TreeStar, Ashland, OR).
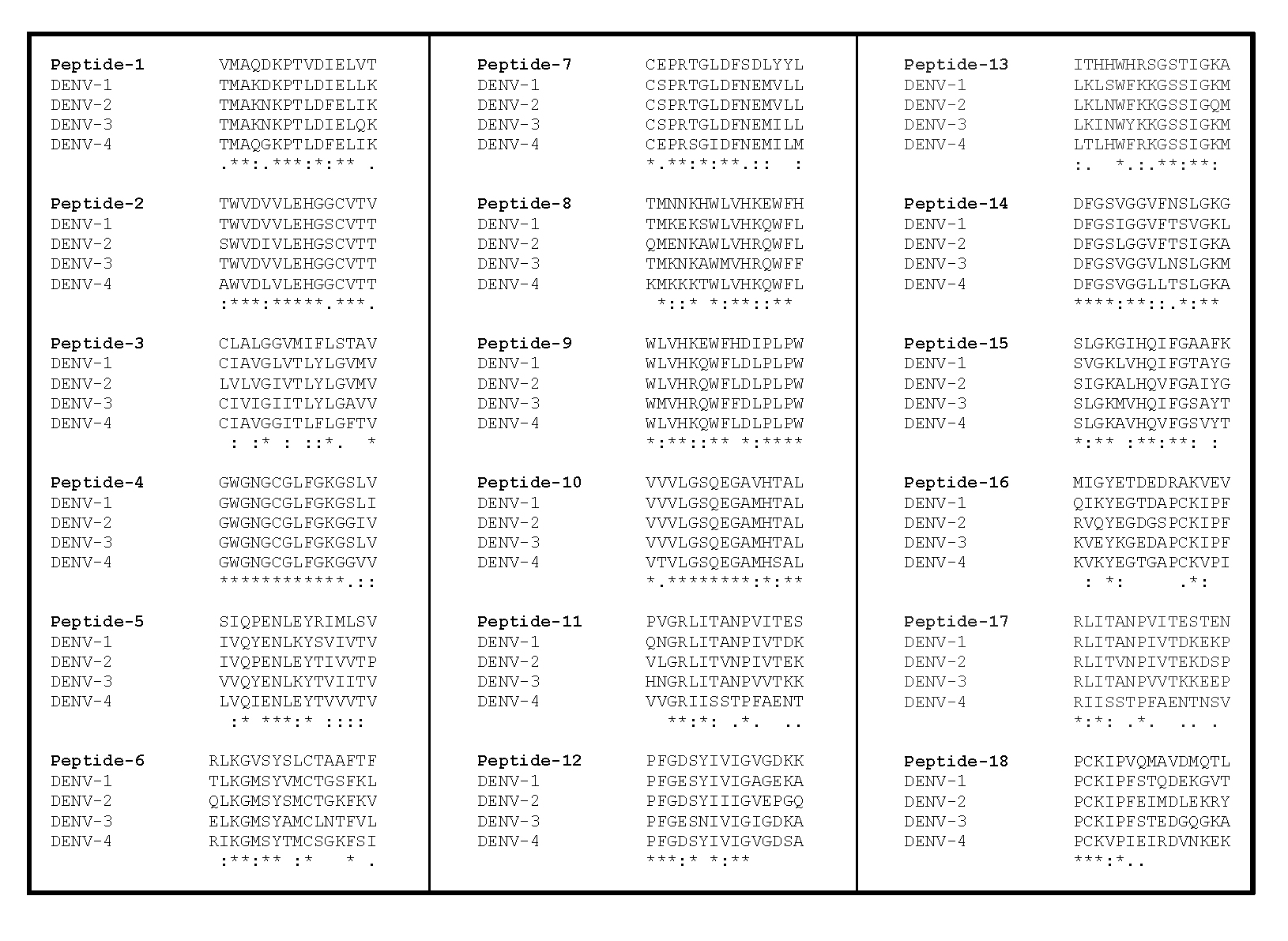
First, we used Net CTL 1.2 [23] with a threshold of 0.75 to predict the HLA-I epitopes from the ZIKV envelope for 3 of the most common alleles in Brazil (HLA-A2 approximately 25%, HLA-A24 approximately 10%, and HLA-B44 approximately 11%) [28, 29]. We identified between 9 and 11 partially overlapping 9-mers for each allele (Table 1). We then proceeded to map these epitopes on an alignment of the ZIKV envelope with the envelopes of DENV 1-4 in UniProt [30]. Based on the similarity of the Net CTL-predicted epitopes between these viruses, we designed 18 peptides (17 peptides were 15-mers and one was a 16-mer) containing at least 1 predicted epitope each (Table 2). Some of the ZIKV peptides had a high similarity with all DENV serotypes (peptides 1, 2, 4, 9, 10, and 14), and some others had a much lower similarity (peptides 3, 16, 17, and 18) (Figure 1).
|
|
|
|
|
Prediction score |
|
|
HLA-A2 |
VMAQDKPTV |
0.6314 |
0.7253 |
0.5250 |
1.0763 |
|
GLFGKGSLV |
0.5499 |
0.9620 |
0.2600 |
0.9770 |
|
|
TMNNKHWLV |
0.6882 |
0.9214 |
0.3690 |
1.1826 |
|
|
RLKGVSYSL |
0.4368 |
0.9783 |
1.2640 |
0.8611 |
|
|
QMAVDMQTL |
0.4571 |
0.7170 |
1.1530 |
0.8466 |
|
|
RLITANPVI |
0.6463 |
0.8378 |
0.8370 |
1.1310 |
|
|
SLGKGIHQI |
0.5049 |
0.9345 |
0.5850 |
0.9220 |
|
|
GMSWFSQIL |
0.5587 |
0.9007 |
0.8630 |
1.0111 |
|
|
QILIGTLLV |
0.5145 |
0.9302 |
0.5280 |
0.9328 |
|
|
ALGGVMIFL |
0.6719 |
0.8577 |
1.0030 |
1.1805 |
|
|
VMIFLSTAV |
0.7082 |
0.9511 |
0.5370 |
1.2252 |
|
|
HLA-A24 |
QYVCKRTLV |
0.2878 |
0.8548 |
0.6800 |
0.7752 |
|
GWGNGCGLF |
0.2630 |
0.4394 |
2.5710 |
0.7544 |
|
|
HWLVHKEWF |
0.4919 |
0.0586 |
2.8090 |
1.1966 |
|
|
WFHDIPLPW |
0.3512 |
0.7479 |
0.8880 |
0.9376 |
|
|
SYSLCTAAF |
0.7168 |
0.8959 |
2.8950 |
1.8055 |
|
|
SLCTAAFTF |
0.3226 |
0.8830 |
2.5640 |
0.9477 |
|
|
PFGDSYIVI |
0.3516 |
0.3935 |
-0.0860 |
0.8034 |
|
|
HWHRSGSTI |
0.5154 |
0.6547 |
0.6970 |
1.2305 |
|
|
DFGSVGGV |
0.3936 |
0.4993 |
2.1670 |
1.0215 |
|
|
HLA-B44 |
LEHGGCVTV |
0.5892 |
0.9765 |
0.2600 |
1.6195 |
|
PENLEYRIM |
0.3196 |
0.2960 |
-0.4540 |
0.8138 |
|
|
LEYRIMLSV |
0.5119 |
0.9701 |
0.5110 |
1.4396 |
|
|
DEDRAKVEV |
0.2847 |
0.7879 |
-0.3030 |
0.8086 |
|
|
CEPRTGLDF |
0.2820 |
0.1832 |
2.2040 |
0.8364 |
|
|
LDFSDLYYL |
0.2548 |
0.7789 |
0.7330 |
0.7849 |
|
|
KEWFHDIPL |
0.7230 |
0.8664 |
1.0750 |
1.9754 |
|
|
QEGAVHTAL |
0.6119 |
0.9682 |
0.7230 |
1.6977 |
|
|
SQILIGTLL |
0.3871 |
0.4819 |
1.0470 |
1.0839 |
Table 2. Selected ZIKV Peptides
|
|
|
|
|
| 1 | VMAQDKPTVDIELVT | 33-47 | A2 |
| 2 | TWVDVVLEHGGCVTV | 19-33 | B44 |
| 3 | CLALGGVMIFLSTAV | 484-498 | A2 |
| 3 | CLALGGVMIFLSTAV | 484-498 | A2 |
| 4 | GWGNGCGLFGKGSLV | 100-114 | A2 |
| 4 | GWGNGCGLFGKGSLV | 100-114 | A24 |
| 5 | SIQPENLEYRIMLSV | 129-143 | B44 |
| 5 | SIQPENLEYRIMLSV | 129-143 | B44 |
| 6 | RLKGVSYSLCTAAFTF | 295-310 | A2 |
| 6 | RLKGVSYSLCTAAFTF | 295-310 | A24 |
| 6 | RLKGVSYSLCTAAFTF | 295-310 | A24 |
| 7 | CEPRTGLDFSDLYYL | 186-200 | B44 |
| 7 | CEPRTGLDFSDLYYL | 186-200 | B44 |
| 8 | TMNNKHWLVHKEWFH | 201-215 | A2 |
| 8 | TMNNKHWLVHKEWFH | 201-215 | A24 |
| 9 | WLVHKEWFHDIPLPW | 207-221 | B44 |
| 9 | WLVHKEWFHDIPLPW | 207-221 | A24 |
| 9 | WLVHKEWFHDIPLPW | 207-221 | A24 |
| 10 | VVVLGSQEGAVHTAL | 251-265 | B44 |
| 11 | PVGRLITANPVITES | 350-365 | A2 |
| 12 | PFGDSYIVIGVGDKK | 377-391 | A24 |
| 13 | ITHHWHRSGSTIGKA | 392-406 | A24 |
| 14 | DFGSVGGVFNSLGKG | 426-440 | A24 |
| 15 | SLGKGIHQIFGAAFK | 436-450 | A2 |
| 16 | MIGYETDEDRAKVEV | 151-165 | B44 |
| 17 | RLITANPVITESTEN | 353-367 | A2 |
| 18 | PCKIPVQMAVDMQTL | 334-348 | A2 |
| A | VMAQDKPTV | 33-41 | A2 |
| B | QDKPTVDIE | 36-44 | - |
| C | PTVDIELVT | 39-47 | - |

To validate the specificity of the peptides, we performed ELISPOTS with PBMCs obtained from 10 control patients from an area free of DENV and ZIKV (New York Blood Bank) and stimulated donor cells with each peptide individually. Stimulation with peptide number 16 resulted in IFNg production above background in 7 out of the 10 donors tested, suggesting that this peptide is cross-reactive with a common antigen. Therefore, we excluded peptide 16 from further analysis. No other peptides induced IFNg production above background levels in the control PBMCs.
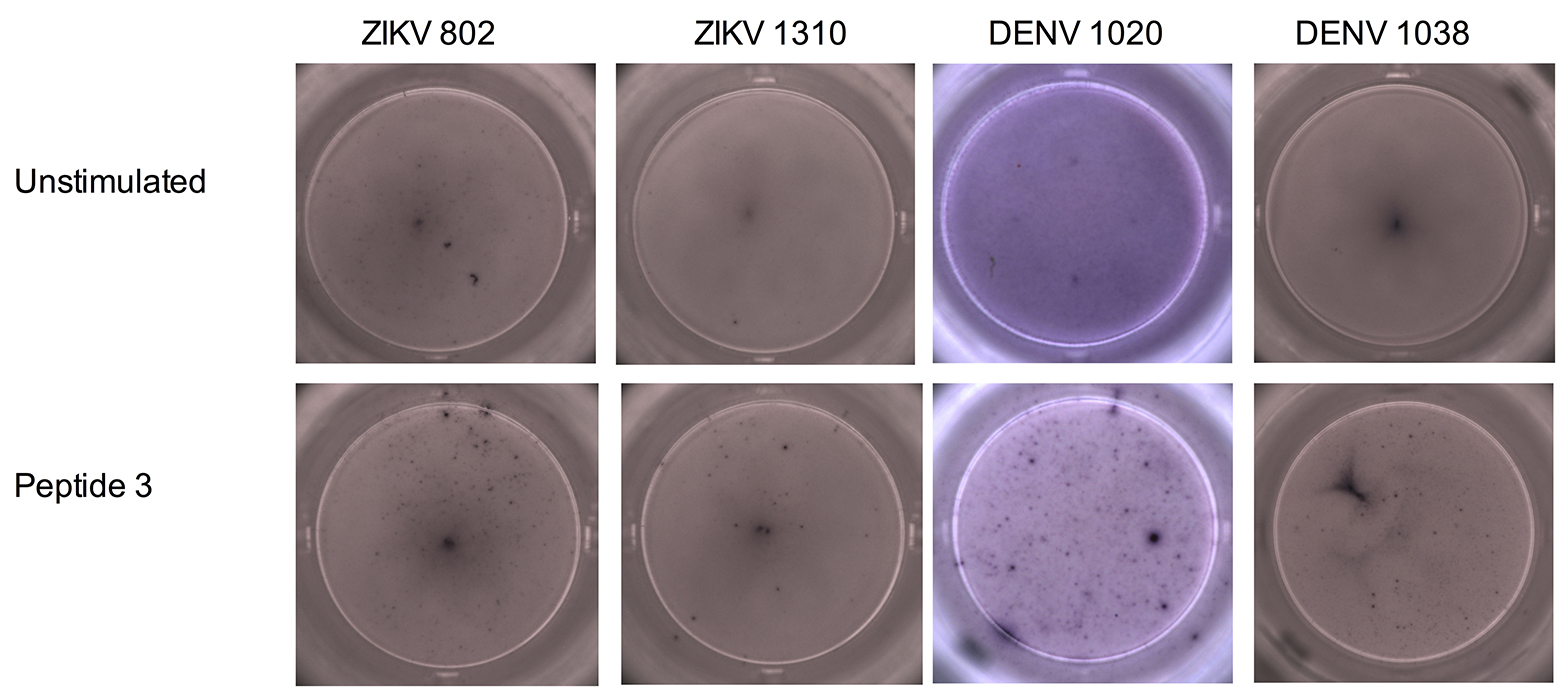
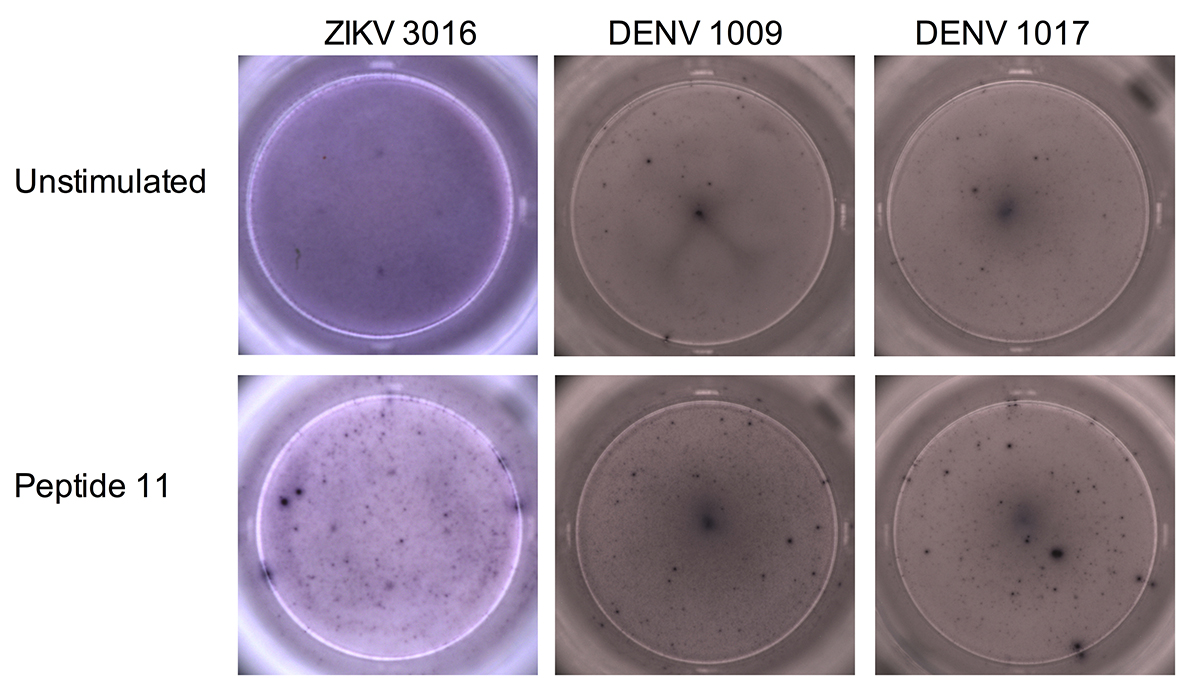
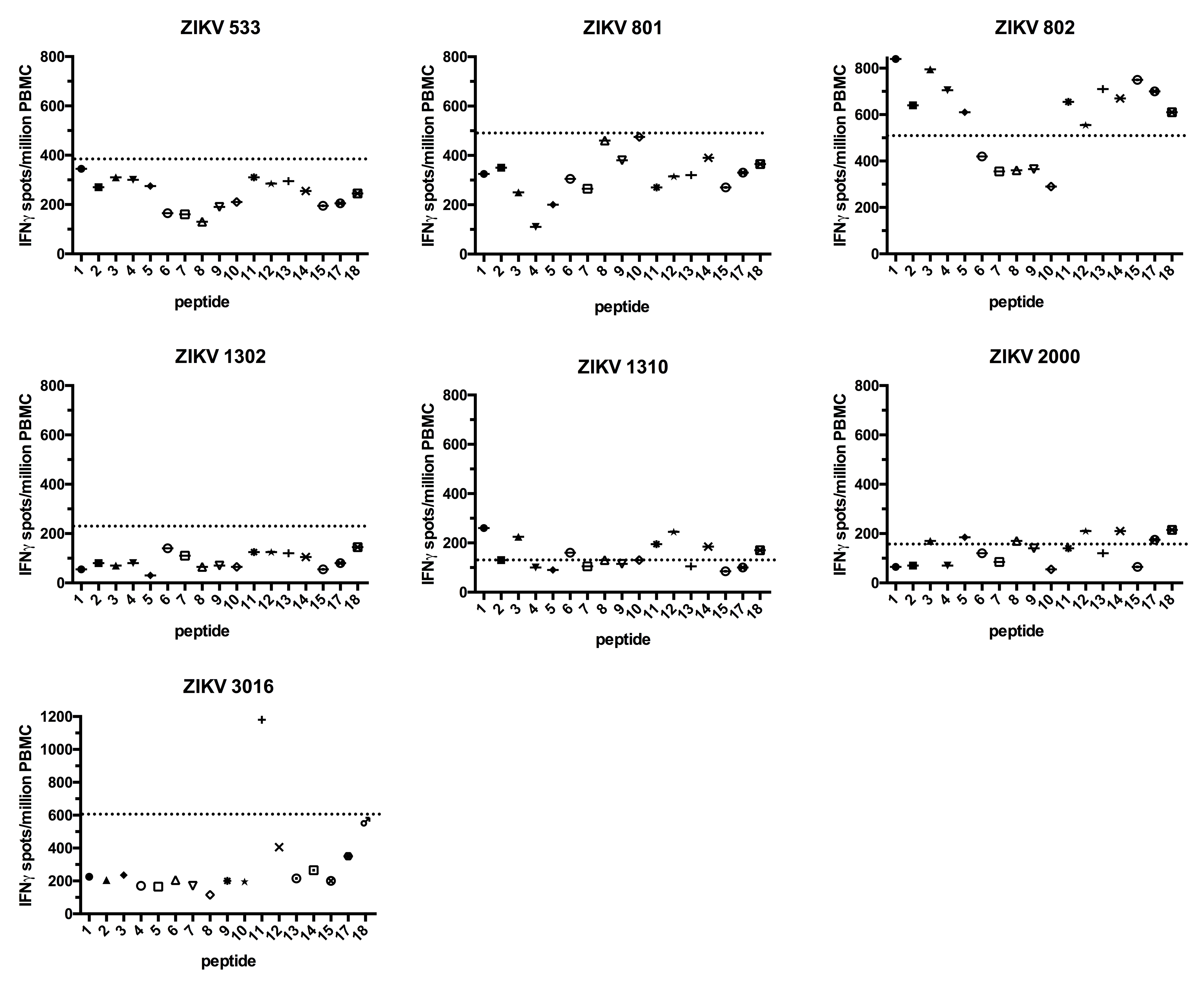
Next, we performed ELISPOTS with PBMCs obtained from 7 Brazilian patients with a previous ZIKV infection (Table 3). The patients denoted ZIKV 533, 801, 1302, and 2000 did not show IFNg production above background for all peptides tested (Figure 2). Patient 3016 responded to peptide 11 only (2-fold over the background), and 802 and 1310 had a weak IFNg response against many peptides including 1, 3, and 11 (less than 2-fold over the background, Figure 2 and Supplementary Figures 1, 2, and 3).
We then stimulated PBMCs obtained from 9 Brazilian individuals, previously vaccinated against DENV (Tables 3 and 4), with the ZIKV peptides. These DENV-vaccinated individuals received the live, attenuated tetravalent vaccine TV003 [31] as part of a phase 2 clinical trial (clinicaltrials.gov identifier: NCT01696422) currently being conducted in Brazil, and samples were collected before ZIKV arrived in São Paulo. Two of the DENV-vaccinated participants (1007 and 1016) did not respond to any of the peptides, and 2 responded only to peptide 11 (1009 and 1017) (Figure 3 and Supplementary Figures 1 and 2). The remaining 5 DENV-vaccinated individuals (1021, 1020, 1038, 1040, and 1049) responded to at least 2 of the peptides, albeit some of the responses were modest (Figure 3 and Supplementary Figures 1 and 3). Overall, peptides 1, 3, and 11 induced a response in both ZIKV-infected and DENV-vaccinated individuals, with the strongest responses observed for peptides 1 and 11. The different time points at which samples were collected made a direct comparison of the magnitude of the IFNg response between the ZIKV-infected patients and the DENV-vaccinated individuals impossible. Some had high background IFNg levels that could have masked weak antigen-specific responses. However, we could still detect antigen-specific responses in those participants (ie, ZIKV 3016, DENV 1009, and DENV 1017).
|
Groups |
Donor ID |
Gender |
Age |
History of Dengue* |
Past YF Immunization* |
Day after the onset of symptoms or DENV-TV003 immunization of blood sample collection |
Reported symptoms |
PCR |
|
|
Urine |
Blood |
||||||||
|
ZIKV |
801 |
F |
53 |
yes (twice) |
yes |
19 |
Fever, myalgia |
+ |
nd |
|
802 |
F |
29 |
no |
yes |
19 |
Myalgia |
+ |
nd |
|
|
533 |
F |
56 |
yes |
yes |
159 |
Myalgia, abdominal pain |
+ |
- |
|
|
1302 |
M |
35 |
no |
yes |
109 |
Myalgia, nausea, abdominal pain |
+ |
nd |
|
|
1310 |
F |
32 |
no |
no |
12 |
Fever, myalgia, retro orbital pain |
nd |
+ |
|
|
2000 ** |
F |
37 |
no |
yes |
15 |
Fever, myalgia, skin rash |
- |
- |
|
|
3016 |
F |
50 |
no |
unk |
16 |
Pruriginous skin rash, conjuctivitis, myalgia |
+ |
+ |
|
|
DENV |
1007 |
F |
38 |
no |
unk |
90 |
Mild skin rash |
na |
na |
|
1009 |
M |
32 |
no |
unk |
90 |
Mild skin rash |
na |
na |
|
|
1011 |
F |
56 |
no |
unk |
90 |
none |
na |
na |
|
|
1016 |
M |
47 |
no |
unk |
90 |
none |
na |
na |
|
|
1017 |
F |
56 |
no |
unk |
90 |
none |
na |
na |
|
|
1020 |
F |
45 |
no |
unk |
90 |
Mild skin rash |
na |
na |
|
|
1021 |
F |
50 |
no |
unk |
90 |
none |
na |
na |
|
|
1038 |
F |
35 |
no |
unk |
90 |
Mild skin rash |
na |
na |
|
|
1040 |
F |
48 |
no |
unk |
90 |
Mild skin rash |
na |
na |
|
|
1049 |
F |
37 |
no |
yes |
90 |
Mild skin rash |
na |
na |
|
| Samples | HLA A | HLA A | HLA B | HLA B | HLA C | HLA C |
| DENV 1007 | A*03:ABWKR | A*29:ABWMY | B*44:ABYSJ | B*58:ACBCJ | C*03:ACPXG | C*16:ACPWC |
| DENV 1009 | A*24:ACPYT | A*24:HYYJ | B*40:ACKGC | B*51:ABUUP | C*03:ABUVB | C*14:ABUWF |
| DENV 1011 | A*02:ZRVU | A*02:ZRVU | B*41:ZRNP | B*50:ABWUY | C*06:ABWZK | C*07:ABWZU |
| DENV 1016 | A*02:ABWJM | A*24:ACAEP | B*44:ABYRU | B*44:ABWUJ | C*05:ACBEA | C*16:ABZVM |
| DENV 1017 | A*29:ABWMX | A*31:ABWNG | B*39:VZGY | B*44:ACBUY | C*07:ABXAG | C*16:ABXBJ |
| DENV 1020 | A*02:ABVHV | A*23:ZKEV | B*27:ACMMT | B*58:ACJEC | C*05:ABZTT | C*07:ABZUE |
| DENV 1021 | A*29:ACDMP | A*29:ACDMP | B*14:ABYJF | B*44:ABZPN | C*08:ACAMF | C*16:ABZVM |
| DENV 1038 | A*02:ABZCH | A*33:ABYFZ | B*15:KPAE | B*44:ACNAV | C*03:ACFEZ | C*14:ACFHD |
| DENV 1040 | A*02:ABUKX | A*29:ABUKV | B*38:WDAZ | B*44:ABZAB | C*12:ABZAC | C*16:ABZAD |
| DENV 1049 | A*02:ABVRS | A*26:ABVSS | B*14:ABWRH | B*51:ABWXE | C*08:ACBFG | C*15:ACBGC |

Finally, we selected peptide 1 to use for fine epitope mapping with PBMCs from participants DENV 1038 and DENV 1049, who were chosen based on their strong responses to this peptide and because they carry the predicted HLA-I allele for this peptide (HLA-A2, Tables 2 and 4). The PBMCs were stimulated with the complete 15-mer peptide or 3 overlapping 9-mer peptides corresponding to the first 9 (A), the middle 9 (B), and the last 9 (C) amino acids of peptide 1 (Table 2). Peptide A was predicted to bind strongly to HLA-A2, and both DENV 1038 and DENV 1049 carried this allele. Stimulation with the complete peptide resulted in an IFNg response, but surprisingly, stimulation with peptide A did not induce a response (Figure 4 A). Similarly, stimulation with peptides B and C resulted in no or weak IFNg responses. The requirement for the full 15-mer peptide suggests that peptide 1 might be presented by HLA-II. To test this, flow cytometry was used to determine if peptide 1 generated a CD4+ or CD8+ T-cell response. The PBMCs from participants DENV 1038 and DENV 1049 were labeled with CellTrace Violet before being cultured for 6 days in the presence or absence of the peptide. The cells were then re-stimulated with the peptide, and antigen-specific cells were identified as CellTrace Violet low and IFNg+. Background IFNg production (CellTrace Violet high, IFNg+) did not change after peptide stimulation, but an antigen- specific response was identified for both donors (Figure 4 B). The majority of the CellTrace Violet low and IFNg+ cells were CD4+ T cells for both DENV-vaccinated participants. We then determined the HLA-II alleles carried by DENV 1038 (HLA-DRB1*04 and HLA-DRB1*13) and DENV 1049 (HLA-DRB1*03 and HLA-DRB1*04) and used the IEDB analysis resource consensus tool [24, 25] to evaluate if peptide 1 could be presented by these alleles. We found that peptide 1 is predicted to be a strong binder for HLA-DRB1*03 (percentile rank 3.22) and HLA-DRB1*13 (percentile rank 2.40). While we only observed a CD4+ T-cell response for subjects DENV 1038 and DENV 1049, it is still possible that other DENV-vaccinated individuals carrying HLA-A2 will generate a CD8+ T-cell response against peptide 1. Because of sample limitations, we could not perform fine epitope mapping for the ZIKV-infected patients. Further studies will be needed to determine the HLA restriction of the other ZIKV epitopes identified.

T-cell cross-reactivity could have a positive or negative impact on ZIKV infection. In fact, many ZIKV-infected people will have had a prior DENV infection or been exposed to other flaviviruses, such as those receiving the YF vaccine. Prior infections or vaccinations sometimes negatively impact the response to a subsequent infection, a phenomenon that has been called “original antigenic sin (OAS)”. This OAS occurs in serial infections with pathogens that share cross-reactive antigens. For example, preformed memory T cells, which cross-react with low avidity to epitopes presented in subsequent infections, could dampen the response of high-avidity T cells [32-34]. Alternatively, cross-reactive T cells might have a protective effect. This is supported by evidence showing that flavivirus-immune individuals have better protection than flavivirus-naive subjects following DENV vaccination [35]; in addition, T-cell cross-reactivity is further complicated by the highly polymorphic nature of HLA molecules. In other words, prior DENV infection or vaccination could impact the ZIKV T-cell response. In mice, ZIKV/DENV cross-reactive CD8+ T cells induced by DENV infection were found to contribute to protection against ZIKV infection [22].
In this study, we have identified ZIKV T-cell epitopes and demonstrated that vaccination against DENV induces a T-cell response against ZIKV. Given the similarity in the T-cell response between DENV natural infection and vaccination [36], it is likely that natural DENV infection can also generate a T-cell response against ZIKV. More research is needed to determine if this cross-reactive immune response is protective or contributory to ZIKV pathology. In particular, the ZIKV CD4+ T-cell response induced by DENV vaccination and identified in this study could contribute to the appearance of cross-reactive antibodies mediating ADE. A better understanding of the interactions between ZIKV and DENV immunity, whether induced by vaccination or by natural infection, will be critical to obtain safe and efficient vaccines.
The authors would like to thank all patients and control participants for their time and efforts towards this study.
The authors declare that no competing interests exist.
Conceived and designed the experiments: DPP FEL CB DFN. Performed the experiments: DPP FEL CGTS AM SKM VDC. Analyzed the data: DPP. Contributed reagents/materials/analysis tools: EGK DFN. Wrote the paper: DPP CB EGK DFN.
This study received partial funding from the Scientific Summit on Zika Virus, held in São Paulo, Brazil on November 3 and 4, 2016 and organized by the George Washington University School of Medicine and Health Sciences: Office of International Medicine Programs. Dominic Paquin-Proulx, Fabio E. Leal, and Douglas F. Nixon are members of the District of Columbia Center for AIDS Research: NIH award number A1117979.
1. Dick GW, Kitchen SF, Haddow AJ. Zika virus. I. Isolations and serological specificity. Trans R Soc Trop Med Hyg. 1952;46(5):509-20. PubMed PMID: 12995440.
2. Driggers RW, Ho CY, Korhonen EM, Kuivanen S, Jaaskelainen AJ, Smura T, Rosenberg A, Hill DA, DeBiasi RL, Vezina G, Timofeev J, Rodriguez FJ, Levanov L, Razak J, Iyengar P, Hennenfent A, Kennedy R, Lanciotti R, du Plessis A, Vapalahti O. Zika Virus Infection with Prolonged Maternal Viremia and Fetal Brain Abnormalities. N Engl J Med. 2016. PubMed PMID: 27028667. doi: 10.1056/NEJMoa1601824
3. Calvet G, Aguiar RS, Melo AS, Sampaio SA, de Filippis I, Fabri A, Araujo ES, de Sequeira PC, de Mendonca MC, de Oliveira L, Tschoeke DA, Schrago CG, Thompson FL, Brasil P, Dos Santos FB, Nogueira RM, Tanuri A, de Filippis AM. Detection and sequencing of Zika virus from amniotic fluid of fetuses with microcephaly in Brazil: a case study. Lancet Infect Dis. 2016. PubMed PMID: 26897108. doi: 10.1016/S1473-3099(16)00095-5
4. Kleber de Oliveira W, Cortez-Escalante J, De Oliveira WT, do Carmo GM, Henriques CM, Coelho GE, Araujo de Franca GV. Increase in Reported Prevalence of Microcephaly in Infants Born to Women Living in Areas with Confirmed Zika Virus Transmission During the First Trimester of Pregnancy - Brazil, 2015. MMWR Morb Mortal Wkly Rep. 2016;65(9):242-7. PubMed PMID: 26963593. doi: 10.15585/mmwr.mm6509e2
5. Martines RB, Bhatnagar J, Keating MK, Silva-Flannery L, Muehlenbachs A, Gary J, Goldsmith C, Hale G, Ritter J, Rollin D, Shieh WJ, Luz KG, Ramos AM, Davi HP, Kleber de Oliveria W, Lanciotti R, Lambert A, Zaki S. Notes from the Field: Evidence of Zika Virus Infection in Brain and Placental Tissues from Two Congenitally Infected Newborns and Two Fetal Losses - Brazil, 2015. MMWR Morb Mortal Wkly Rep. 2016;65(6):159-60. PubMed PMID: 26890059. doi: 10.15585/mmwr.mm6506e1
6. Mlakar J, Korva M, Tul N, Popovic M, Poljsak-Prijatelj M, Mraz J, Kolenc M, Resman Rus K, Vesnaver Vipotnik T, Fabjan Vodusek V, Vizjak A, Pizem J, Petrovec M, Avsic Zupanc T. Zika Virus Associated with Microcephaly. N Engl J Med. 2016;374(10):951-8. PubMed PMID: 26862926. doi: 10.1056/NEJMoa1600651
7. Cugola FR, Fernandes IR, Russo FB, Freitas BC, Dias JL, Guimaraes KP, Benazzato C, Almeida N, Pignatari GC, Romero S, Polonio CM, Cunha I, Freitas CL, Brandao WN, Rossato C, Andrade DG, Faria Dde P, Garcez AT, Buchpigel CA, Braconi CT, Mendes E, Sall AA, Zanotto PM, Peron JP, Muotri AR, Beltrao-Braga PC. The Brazilian Zika virus strain causes birth defects in experimental models. Nature. 2016;534(7606):267-71. PubMed PMID: 27279226. Pubmed Central PMCID: PMC4902174. doi: 10.1038/nature18296
8. Yockey LJ, Varela L, Rakib T, Khoury-Hanold W, Fink SL, Stutz B, Szigeti-Buck K, Van den Pol A, Lindenbach BD, Horvath TL, Iwasaki A. Vaginal Exposure to Zika Virus during Pregnancy Leads to Fetal Brain Infection. Cell. 2016;166(5):1247-56 e4. PubMed PMID: 27565347. Pubmed Central PMCID: PMC5006689. doi: 10.1016/j.cell.2016.08.004
9. Adams Waldorf KM, Stencel-Baerenwald JE, Kapur RP, Studholme C, Boldenow E, Vornhagen J, Baldessari A, Dighe MK, Thiel J, Merillat S, Armistead B, Tisoncik-Go J, Green RR, Davis MA, Dewey EC, Fairgrieve MR, Gatenby JC, Richards T, Garden GA, Diamond MS, Juul SE, Grant RF, Kuller L, Shaw DW, Ogle J, Gough GM, Lee W, English C, Hevner RF, Dobyns WB, Gale M, Jr., Rajagopal L. Fetal brain lesions after subcutaneous inoculation of Zika virus in a pregnant nonhuman primate. Nat Med. 2016;22(11):1256-9. PubMed PMID: 27618651. doi: 10.1038/nm.4193
10. Cauchemez S, Besnard M, Bompard P, Dub T, Guillemette-Artur P, Eyrolle-Guignot D, Salje H, Van Kerkhove MD, Abadie V, Garel C, Fontanet A, Mallet HP. Association between Zika virus and microcephaly in French Polynesia, 2013-15: a retrospective study. Lancet. 2016. PubMed PMID: 26993883. doi: 10.1016/S0140-6736(16)00651-6
11. Brasil P, Pereira JP, Jr., Raja Gabaglia C, Damasceno L, Wakimoto M, Ribeiro Nogueira RM, Carvalho de Sequeira P, Machado Siqueira A, Abreu de Carvalho LM, Cotrim da Cunha D, Calvet GA, Neves ES, Moreira ME, Rodrigues Baiao AE, Nassar de Carvalho PR, Janzen C, Valderramos SG, Cherry JD, Bispo de Filippis AM, Nielsen-Saines K. Zika Virus Infection in Pregnant Women in Rio de Janeiro - Preliminary Report. N Engl J Med. 2016. PubMed PMID: 26943629. doi: 10.1056/NEJMoa1602412
12. Johansson MA, Mier YT-RL, Reefhuis J, Gilboa SM, Hills SL. Zika and the Risk of Microcephaly. N Engl J Med. 2016. PubMed PMID: 27222919. doi: 10.1056/NEJMp1605367
13. Brasil P, Pereira JP, Jr., Moreira ME, Ribeiro Nogueira RM, Damasceno L, Wakimoto M, Rabello RS, Valderramos SG, Halai UA, Salles TS, Zin AA, Horovitz D, Daltro P, Boechat M, Raja Gabaglia C, Carvalho de Sequeira P, Pilotto JH, Medialdea-Carrera R, Cotrim da Cunha D, Abreu de Carvalho LM, Pone M, Machado Siqueira A, Calvet GA, Rodrigues Baiao AE, Neves ES, Nassar de Carvalho PR, Hasue RH, Marschik PB, Einspieler C, Janzen C, Cherry JD, Bispo de Filippis AM, Nielsen-Saines K. Zika Virus Infection in Pregnant Women in Rio de Janeiro. N Engl J Med. 2016;375(24):2321-34. PubMed PMID: 26943629. doi: 10.1056/NEJMoa1602412
14. Honein MA, Dawson AL, Petersen EE, Jones AM, Lee EH, Yazdy MM, Ahmad N, Macdonald J, Evert N, Bingham A, Ellington SR, Shapiro-Mendoza CK, Oduyebo T, Fine AD, Brown CM, Sommer JN, Gupta J, Cavicchia P, Slavinski S, White JL, Owen SM, Petersen LR, Boyle C, Meaney-Delman D, Jamieson DJ, Collaboration USZPR. Birth Defects Among Fetuses and Infants of US Women With Evidence of Possible Zika Virus Infection During Pregnancy. JAMA. 2017;317(1):59-68. PubMed PMID: 27960197. doi: 10.1001/jama.2016.19006
15. Cuevas EL, Tong VT, Rozo N, Valencia D, Pacheco O, Gilboa SM, Mercado M, Renquist CM, Gonzalez M, Ailes EC, Duarte C, Godoshian V, Sancken CL, Turca AM, Calles DL, Ayala M, Morgan P, Perez EN, Bonilla HQ, Gomez RC, Estupinan AC, Gunturiz ML, Meaney-Delman D, Jamieson DJ, Honein MA, Martinez ML. Preliminary Report of Microcephaly Potentially Associated with Zika Virus Infection During Pregnancy - Colombia, January-November 2016. MMWR Morb Mortal Wkly Rep. 2016;65(49):1409-13. PubMed PMID: 27977645. doi: 10.15585/mmwr.mm6549e1
16. Dejnirattisai W, Supasa P, Wongwiwat W, Rouvinski A, Barba-Spaeth G, Duangchinda T, Sakuntabhai A, Cao-Lormeau VM, Malasit P, Rey FA, Mongkolsapaya J, Screaton GR. Dengue virus sero-cross-reactivity drives antibody-dependent enhancement of infection with zika virus. Nat Immunol. 2016. PubMed PMID: 27339099. doi: 10.1038/ni.3515
17. Priyamvada L, Quicke KM, Hudson WH, Onlamoon N, Sewatanon J, Edupuganti S, Pattanapanyasat K, Chokephaibulkit K, Mulligan MJ, Wilson PC, Ahmed R, Suthar MS, Wrammert J. Human antibody responses after dengue virus infection are highly cross-reactive to Zika virus. Proc Natl Acad Sci U S A. 2016. PubMed PMID: 27354515. doi: 10.1073/pnas.1607931113
18. Castanha PM, Nascimento EJ, Cynthia B, Cordeiro MT, de Carvalho OV, de Mendonca LR, Azevedo EA, Franca RF, Rafael D, Marques ET, Jr. Dengue virus (DENV)-specific antibodies enhance Brazilian Zika virus (ZIKV) infection. J Infect Dis. 2016. PubMed PMID: 28039355. doi: 10.1093/infdis/jiw638
19. Paul LM, Carlin ER, Jenkins MM, Tan AL, Barcellona CM, Nicholson CO, Michael SF, Isern S. Dengue virus antibodies enhance Zika virus infection. Clin Transl Immunology. 2016;5(12):e117. PubMed PMID: 28090318. Pubmed Central PMCID: PMC5192063. doi: 10.1038/cti.2016.72
20. Halstead SB. Biologic Evidence Required for Zika Disease Enhancement by Dengue Antibodies. Emerg Infect Dis. 2017;23(4):569-73. PubMed PMID: 28322690. doi: 10.3201/eid2304.161879
21. Weiskopf D, Cerpas C, Angelo MA, Bangs DJ, Sidney J, Paul S, Peters B, Sanches FP, Silvera CG, Costa PR, Kallas EG, Gresh L, de Silva AD, Balmaseda A, Harris E, Sette A. Human CD8+ T-Cell Responses Against the 4 Dengue Virus Serotypes Are Associated With Distinct Patterns of Protein Targets. J Infect Dis. 2015;212(11):1743-51. PubMed PMID: 25980035. Pubmed Central PMCID: PMC4633759. doi: 10.1093/infdis/jiv289
22. Wen J, Tang WW, Sheets N, Ellison J, Sette A, Kim K, Shresta S. Identification of Zika virus epitopes reveals immunodominant and protective roles for dengue virus cross-reactive CD8+ T cells. Nat Microbiol. 2017;2:17036. PubMed PMID: 28288094. doi: 10.1038/nmicrobiol.2017.36
23. Larsen MV, Lundegaard C, Lamberth K, Buus S, Lund O, Nielsen M. Large-scale validation of methods for cytotoxic T-lymphocyte epitope prediction. BMC Bioinformatics. 2007;8:424. PubMed PMID: 17973982. Pubmed Central PMCID: PMC2194739. doi: 10.1186/1471-2105-8-424
24. Wang P, Sidney J, Dow C, Mothe B, Sette A, Peters B. A systematic assessment of MHC class II peptide binding predictions and evaluation of a consensus approach. PLoS Comput Biol. 2008;4(4):e1000048. PubMed PMID: 18389056. Pubmed Central PMCID: PMC2267221. doi: 10.1371/journal.pcbi.1000048
25. Wang P, Sidney J, Kim Y, Sette A, Lund O, Nielsen M, Peters B. Peptide binding predictions for HLA DR, DP and DQ molecules. BMC Bioinformatics. 2010;11:568. PubMed PMID: 21092157. Pubmed Central PMCID: PMC2998531. doi: 10.1186/1471-2105-11-568
26. Li W, Cowley A, Uludag M, Gur T, McWilliam H, Squizzato S, Park YM, Buso N, Lopez R. The EMBL-EBI bioinformatics web and programmatic tools framework. Nucleic Acids Res. 2015;43(W1):W580-4. PubMed PMID: 25845596. Pubmed Central PMCID: PMC4489272. doi: 10.1093/nar/gkv279
27. McWilliam H, Li W, Uludag M, Squizzato S, Park YM, Buso N, Cowley AP, Lopez R. Analysis Tool Web Services from the EMBL-EBI. Nucleic Acids Res. 2013;41(Web Server issue):W597-600. PubMed PMID: 23671338. Pubmed Central PMCID: PMC3692137. doi: 10.1093/nar/gkt376
28. Bardi MS, Jarduli LR, Jorge AJ, Camargo RB, Carneiro FP, Gelinski JR, Silva RA, Lavado EL. HLA-A, B and DRB1 allele and haplotype frequencies in volunteer bone marrow donors from the north of Parana State. Rev Bras Hematol Hemoter. 2012;34(1):25-30. PubMed PMID: 23049380. Pubmed Central PMCID: PMC3459602. doi: 10.5581/1516-8484.20120010
29. Ayo CM, da Silveira Camargo AV, Xavier DH, Batista MF, Carneiro OA, Brandao de Mattos CC, Ricci O, Jr., de Mattos LC. Frequencies of allele groups HLA-A, HLA-B and HLA-DRB1 in a population from the northwestern region of Sao Paulo State, Brazil. Int J Immunogenet. 2015;42(1):19-25. PubMed PMID: 25418108. doi: 10.1111/iji.12159
30. UniProt C. UniProt: a hub for protein information. Nucleic Acids Res. 2015;43(Database issue):D204-12. PubMed PMID: 25348405. Pubmed Central PMCID: PMC4384041. doi: 10.1093/nar/gku989
31. Kirkpatrick BD, Whitehead SS, Pierce KK, Tibery CM, Grier PL, Hynes NA, Larsson CJ, Sabundayo BP, Talaat KR, Janiak A, Carmolli MP, Luke CJ, Diehl SA, Durbin AP. The live attenuated dengue vaccine TV003 elicits complete protection against dengue in a human challenge model. Sci Transl Med. 2016;8(330):330ra36. PubMed PMID: 27089205. doi: 10.1126/scitranslmed.aaf1517
32. Zehn D, Turner MJ, Lefrancois L, Bevan MJ. Lack of original antigenic sin in recall CD8(+) T cell responses. J Immunol. 2010;184(11):6320-6. PubMed PMID: 20439913. Pubmed Central PMCID: PMC2982183. doi: 10.4049/jimmunol.1000149
33. Mongkolsapaya J, Dejnirattisai W, Xu XN, Vasanawathana S, Tangthawornchaikul N, Chairunsri A, Sawasdivorn S, Duangchinda T, Dong T, Rowland-Jones S, Yenchitsomanus PT, McMichael A, Malasit P, Screaton G. Original antigenic sin and apoptosis in the pathogenesis of dengue hemorrhagic fever. Nat Med. 2003;9(7):921-7. PubMed PMID: 12808447. doi: 10.1038/nm887
34. Rivino L, Lim MQ. CD4+ and CD8+ T-cell immunity to Dengue - lessons for the study of Zika virus. Immunology. 2017;150(2):146-54. PubMed PMID: 27763656. Pubmed Central PMCID: PMC5214511. doi: 10.1111/imm.12681
35. Hadinegoro SR, Arredondo-Garcia JL, Capeding MR, Deseda C, Chotpitayasunondh T, Dietze R, Muhammad Ismail HI, Reynales H, Limkittikul K, Rivera-Medina DM, Tran HN, Bouckenooghe A, Chansinghakul D, Cortes M, Fanouillere K, Forrat R, Frago C, Gailhardou S, Jackson N, Noriega F, Plennevaux E, Wartel TA, Zambrano B, Saville M, Group C-TDVW. Efficacy and Long-Term Safety of a Dengue Vaccine in Regions of Endemic Disease. N Engl J Med. 2015;373(13):1195-206. PubMed PMID: 26214039. doi: 10.1056/NEJMoa1506223
36. Angelo MA, Grifoni A, O'Rourke PH, Sidney J, Paul S, Peters B, de Silva AD, Phillips E, Mallal S, Diehl SA, Kirkpatrick BD, Whitehead SS, Durbin AP, Sette A, Weiskopf D. Human CD4+ T Cell Responses to an Attenuated Tetravalent Dengue Vaccine Parallel Those Induced by Natural Infection in Magnitude, HLA Restriction, and Antigen Specificity. J Virol. 2017;91(5). PubMed PMID: 27974563. Pubmed Central PMCID: PMC5309943. doi: 10.1128/JVI.02147-16