Gabrielle N. Gaultier1, Kayla N. Colledanchise1, Alaa Alhazmi1, Marina Ulanova1,2
Background: We have recently found that lipooligosaccharide (LOS) isolated from encapsulated strains of Haemophilus influenzae (H. influenzae) has strong adjuvant, but diminished pro-inflammatory ability as compared to Escherichia coli lipopolysaccharide (LPS). In this study, we aimed to determine the immunostimulatory capacity of nontypeable/ non-encapsulated H. influenzae (NTHi) LOS by comparing the effect of killed bacteria with LOS isolated from the same strain.
Methods: Following stimulation of human monocytic THP-1 cells with killed NTHi strain 375, or with the corresponding amount of LOS, we studied the protein and gene expression of immunostimulatory and antigen-presenting molecules, cytokines, and innate immune receptors.
Results: Stimulation
with LOS resulted in lower expression of adhesion (CD54, CD58) as well as
costimulatory molecules (CD40, CD86), but in higher expression of
antigen-presenting molecules (HLA-DR and HLA-ABC) compared to killed NTHi,
whereas killed bacteria induced higher release of both TNF-
Conclusions: Considering the important role of NTHi as a respiratory pathogen, and its currently increasing significance in the etiology of invasive infections, LOS deserves further attention as a vaccine antigen, which also has potent adjuvant properties.
Keywords: Nontypeable Haemophilus influenzae, Lipooligosaccharide, THP-1 cells, innate immune responses
Nonencapsulated, or nontypeable Haemophilus influenzae (NTHi) is an important gram-negative human pathogen, which frequently colonizes the upper respiratory tract and is a common cause of local infections, such as sinusitis, pediatric otitis media, or exacerbations of chronic obstructive pulmonary disease (COPD) [1, 2]. During the last decade, an increasing incidence of invasive diseases caused by NTHi, including sepsis, meningitis, or bacteremic pneumonia has been reported worldwide, especially in newborns, immunocompromised individuals, and the elderly [3-5]. No specific vaccine to prevent NTHi infections is currently available, although a common H. influenzae antigen, protein D is included in the pediatric vaccine Synflorix® as a carrier protein for pneumococcal polysaccharide antigens. Clinical trials demonstrated that this vaccine was 35% efficient in preventing otitis media caused by NTHi in young children; however, it remains unknown if Synflorix® can offer protection against invasive NTHi disease [6]. Several H. influenzae antigenic compounds have been considered as potential NTHi-vaccine candidates, and some have been tested in animal models; however, none went beyond phase I clinical trials [7-12].
Lipooligosaccharide (LOS) is a major virulence factor of H. influenzae that is responsible for inflammatory responses associated with this infection [13]. This LOS is similar to one of the most studied endotoxin molecules, the lipopolysaccharide (LPS) of gram-negative bacteria. The structure of LPS consists of 3 covalently -linked components: lipid A embedded into the outer membrane, the oligosaccharide core, and the polysaccharide or O-antigen that covers the surface of the bacteria [14], and LPS activates the innate immune response once bound to the cellular receptor CD14, myeloid differentiation protein 2 (MD-2), and Toll-like receptor 4 (TLR4) [15]. Lipid A is the major pathogen-associated molecular pattern recognized by the innate immune system; the number of acyl chains determines the level of response that occurs [15]. In an in vivo situation, LPS causes a strong activation of the innate immune system that results in a massive production of pro-inflammatory cytokines clinically associated with the development of septic shock [14]. Haemophilus influenzae LOS has a similar lipid A structure and function to LPS, but it lacks repeating units of O-antigen side chain. The structure of LOS consists of an oligosaccharide core, which is made up of a triheptose that is linked to a 3-deoxy-D-manno-oct-2-ulosonic acid (Kdo) residue through covalent bonds. The core region is also covalently linked to the lipid A component, as well as an acylated glucosamine disaccharide backbone that is anchored to the outer membrane of bacteria [16, 17].
Our recent studies demonstrated that LOS isolated from either an encapsulated H. influenzae serotype b strain (Eagan), or a capsule-deficient serotype d strain (Rd) had diminished capacity to induce pro-inflammatory responses in comparison to a prototypic endotoxin of gram-negative bacteria, Escherichia coli LPS. However, LOS and LPS had comparable abilities to up-regulate the expression of co-stimulatory and antigen-presenting molecules [18]. Moreover, we have found the presence of naturally acquired bactericidal antibodies against NTHi LOS in sera of healthy adults suggesting their protective role against invasive disease [19]. Taking into consideration that LOS represents a common component of all H. influenzae strains and combines antigenic and adjuvant properties, it deserves further attention as a plausible vaccine candidate to protect against NTHi infections. In this study, we sought to elucidate the role of LOS in the overall immunostimulatory capacities of NTHi by comparing the effect of killed NTHi bacteria with the effect of LOS isolated from the same strain.
The human THP-1 monocytic leukemia cell line (ATCC, TIB-202) was stored in liquid nitrogen until thawed for culturing and maintained as described by Choi et al 2014 [18]. Cell number and viability were determined with a Bright-Line Hemacytometer (Hausser Scientific, Horsham, PA) using a 1:1 dilution factor with 0.4% Trypan blue solution (Sigma- Aldrich, St. Louis, MO). To induce differentiation, THP-1 cells were plated at 1 × 106 cells/1 mL/well in 24-well flat-bottom plates (Costar, Corning Incorporated, Corning NY), in RPMI 1640 medium (Sigma-Aldrich) supplemented with 10% heat-inactivated fetal bovine serum (SAFC Biosciences, Lenexa, KS) and 200 μl antibiotic-antimycotic (complete medium) (Life Technologies, Inc., Burlington, Ontario). Cells were then treated with 20 ng/mL phorbol myristate acetate (PMA; Sigma-Aldrich) at 37°C in 5% CO2 for 12 hours, then washed and re-suspended in the same medium. After 48 hours of further incubation, the cells were washed twice with serum- and antibiotic-free medium and used for experiments.
NTHi strain 375, originally isolated from a pediatric patient with otitis media [20], was grown on brain-heart infusion (BHI) plates supplemented with 10 mg/mL hemin and 1 μg/mL nicotine adenine dinucleotide (NAD) overnight, in 5% CO2 at 37°C. Bacteria were then transferred to supplemented BHI broth and grown to log phase at 37°C with shaking. To kill bacteria, a NTHi suspension with an optical density (OD) of 0.1 read at 600 nm was prepared and treated with 100 μg/mL gentamicin (Sigma-Aldrich) for 30 minutes at 37°C with shaking. Complete killing was confirmed using drop plating. Bacteria were washed twice with phosphate buffered saline (PBS), resuspended in PBS, and used for stimulation of THP-1 cells. The LOS from NTHi 375 was isolated and purified using proteinase K as described by Choi et al [18].
In order to estimate the number of bacteria that contain certain amounts of LOS, previously reported values of cell-bound LOS were used. Gu et al determined the range of cell-bound LOS per bacterium for NTHi strains 9274, 6491, 5756, 2627, and 2019 as 1.6 × 106 to 4.8 x 106 molecules [21]. For our calculations, we considered an average of the lowest and highest reported values, ie, 3.2 × 106 molecules of LOS per bacterium. Using this value and Avogadro’s number (6.022 × 1023), the moles of LOS per bacterium were determined. As the molecular weight of LOS has been estimated as 4,000 g/mol, based on known structures of oligosaccharides and lipid A from H. influenzae LOS [21], it was multiplied by the moles of LOS per bacterium to obtain an average weight of LOS per bacterium and used to determine the number of bacteria harboring certain amounts of LOS as indicated in Table 1. The numbers of bacteria used for experiments were divided by the number of THP-1 cells to determine the approximate NTHi to THP-1 cell ratio (Table 1).
Table 1. Estimated numbers of bacteria corresponding to LOS doses used in this study. * Gene expression analysis (qPCR), ** Surface molecule expression (flow cytometry and ELISA analysis)
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
A total of 0.5 × 106 THP-1 cells in 2 mL of complete medium were placed in each well of a 12-well plate (Costar) for 24 hours. Cells were stimulated with 1 mg/mL of E. coli 0111:B4 LPS (Sigma-Aldrich), or 1, 5, 10, or 15 mg/mL NTHi 375 LOS, or the corresponding amounts of killed bacteria for 24 hours as described by Choi et al [18]. Following stimulation, cells were washed once with PBS and immunostained with fluorochrome- conjugated antibodies against CD54 (ICAM-1), CD40, CD86 (B7-2) (BD Biosciences, Mississauga, Ontario), CD58 (LFA-3) (Cedarlane, Burlington, Ontario), HLA-DR (major histocompatibility complex (MHC) class II) (Biolegend, San Diego, CA) HLA-ABC (MHC class I) antibody (BD Biosciences) or the corresponding isotype control as was previously described [ 18].
The experiments were performed as previously described [18]. Briefly, 1.5 × 106 THP-1 cells were suspended in 2 mL
complete medium and the concentrations of 0.1 and 1 mg/mL of NTHi 375 LOS,
or E. coli LPS were used to stimulate the cells for 4 hours,
followed by RNA extraction and qPCR analysis. Gene expression of TNF-
Differentiated THP-1 cells were stimulated with NTHi LOS at concentrations
of 1, 5, 10, and 15 µg/ml, or corresponding amounts of killed bacteria, or E. coli 0111:B4 LPS at concentration of 1 mg/mL for 24 hours. Cell
culture supernatants were aliquoted and stored at −80°C for cytokine
measurement. The concentrations of IL-1β, TNF-
Data were expressed as a mean of at least 3 independent experiments. Statistical significance was determined with a 1-way analysis of variance (ANOVA) with Newman-Keuls multiple comparison post hoc test, where P values less than 0.05 were considered significant (Prism, GraphPad version 5.0, La Jolla, CA).
To study the ability of LOS to activate innate immune responses, THP-1 cells were stimulated with either NTHi 375 LOS, or a potent innate immune activator E. coli 0111:B4 LPS, and cell surface expression of CD54 (ICAM-1), CD40, CD58 (LFA-1), CD86 (B7), HLA- ABC (MHC class I), and HLA-DR (MHC class II) was assessed using flow cytometry analysis. As a negative control, unstimulated THP-1 cells were used. Although stimulation with NTHi LOS caused upregulation of CD54, CD40, and CD58 expression in THP-1 cells, this effect was significantly reduced compared to stimulation with the same dose of E. coli LPS, ie 1 mg/mL (Figure 1A, C, D). However, NTHi LOS and E. coli LPS induced a similar increase in the expression of both MHC class I and II molecules (Figure 1E, F). Unlike the effect of LPS, stimulation of THP-1 cells with 1 mg/mL of NTHi LOS did not cause an increase in the expression of CD86 (Figure 1B).
Figure 1. Flow cytometry analysis of the expression of co-stimulatory and antigen-presenting molecules in THP-1 cells stimulated with NTHi LOS or E. coli LPS. THP-1 cells were stimulated with 1.0 mg/ml of LPS or LOS for 24 hours. Unstimulated THP-1 cells (0.5 × 106) in complete culture medium served as a negative control. Cell surface expression of CD54 (A), CD86 (B), CD40 (C), CD58 (D) HLA-ABC (E), and HLA-DR (F) was measured and data expressed as the mean fluorescence intensity (MFI); the bars indicate the mean with the standard error of the mean (SEM) ( n=3 ). *, P < 0.05 compared to the negative control; ·, P < 0.05 between LPS and LOS.
These results indicate that although LOS of NTHi 375 shows decreased
ability to induce the surface expression of co-stimulatory molecules CD54,
CD40, CD58, and CD86 as compared to the same dose of LPS, it has a similar
ability to up-regulate the expression of antigen-presenting molecules MHC
class I and II. Furthermore, LOS has decreased ability to induce gene
expression of pro-inflammatory cytokines TNF-
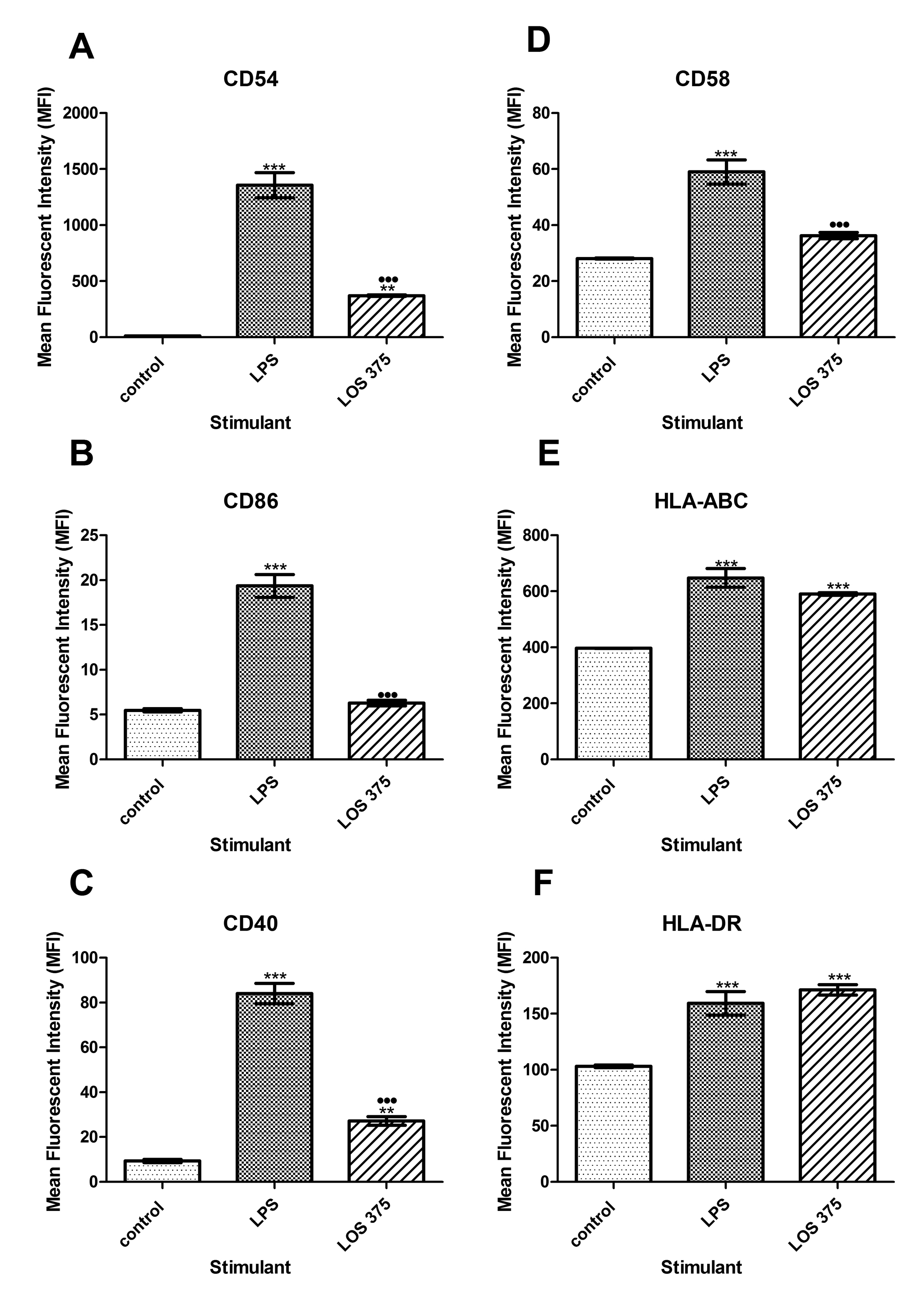
To examine the contribution of NTHi LOS to the immunostimulatory ability of whole bacteria we compared the results of the exposure of THP-1 cells to increasing doses of LOS with the corresponding numbers of killed NTHi of the same strain. In comparison to the effect of killed NTHi, LOS stimulation resulted in lower expression of CD54, CD40, and CD86 (at doses between 1 and 15 mg/mL), and of CD58 (at doses of 1 and 5 mg/mL) (Figure 3A-D) suggesting that in addition to LOS, other immunostimulatory bacterial components may contribute to the upregulation of these surface molecules. The effect of LOS was dose-dependent, and in the case of CD86, the highest LOS dose, (15 mg/mL), induced the expression of this co-stimulatory molecule in a manner comparable to LPS (Figure 3B).
Figure 3. Flow cytometry analysis of the expression of co-stimulatory and antigen-presenting molecules in THP-1 cells stimulated with E. coli LPS, NTHi LOS or killed NTHi. THP-1 cells were stimulated with 1.0 mg/ml of LPS, or LOS at concentrations of 1, 5, 10, and 15 mg/ml, or corresponding amounts of killed NTHi: 4.762 × 107, 2.381 × 108, 4.762 × 108, 7.143 × 108 CFU per 0.5 × 106 THP-1 cells for 24 hours. Unstimulated THP-1 cells (0.5 × 106) in complete culture medium served as a negative control. Cell surface expression of CD54 (A), CD86 (B), CD40 (C), CD58 (D) HLA-ABC (E), and HLA-DR (F) was measured and data expressed as the mean fluorescence intensity (MFI) with the standard error of the mean (SEM) (n = 3). *, P < 0.05 compared to the unstimulated control; +, P < 0.05 between LOS and killed NTHi; ·, P < 0.05 between LPS and LOS. (G): Original histograms of 1 representative experiment showing HLA-DR expression in THP-1 cells stimulated with 10 mg/ml of NTHi LOS (dark grey), or with the corresponding amounts of killed NTHi (open histogram); light grey: unstimulated cells stained with the isotype control antibody.Interestingly, LOS induced significantly higher expression of antigen-presenting molecules compared to the effect of the corresponding numbers of whole bacteria. For HLA-DR, this effect was consistent for all the doses of LOS; for HLA-ABC, the effect only appeared at the highest LPS concentration (15 mg/mL) (Figure 3E-G). Because the cellular expression of MHC molecules is largely regulated by a balance of the cytokines IFN-γ and IL-10, we studied their release by differentiated THP-1 cells stimulated with purified LOS or killed NTHi.
To study the release of cytokines TNF-
Clinical infections caused by NTHi are characterized by intense
inflammation that has been largely attributed to LOS, the major component
of the H. influenzae cell wall [22]; LOS has the capacity to activate pro-inflammatory responses due to the
interaction of lipid A with TLR4-signaling complex resulting in NF-κB
activation with subsequent expression of multiple pro-inflammatory
molecules [23]. Indeed, in our model, we observed that exposure of human monocytic cells
to killed NTHi in vitro resulted in a large release of TNF-
Other NTHi molecules that can potentially be recognized by innate immune receptors and are capable of inducing inflammatory responses include lipopeptides and porins (TLR2 ligands), and bacterial DNA (TLR9 ligand) [29-33]. Moreover, a cross talk among different pattern recognition receptors activated by several microbial products can result in the amplification of cellular responses [23]. Recent studies indicated that NTHi could activate the NLRP3 inflammasome [34]. Indeed, we found that both whole NTHi and isolated LOS induced release of mature IL-1β that requires inflammasome activation, in addition to the transcription of the IL-1β gene, which is largely regulated by NF-κB [35].
The results of this study corroborate our earlier observations that the pro-inflammatory ability of H. influenzae LOS is significantly decreased in comparison to E. coli LPS, the prototypic endotoxin of gram-negative bacteria [18]. In particular, the ability of LOS isolated from H. influenzae serotype b strain Eagan and from the serotype d strain Rd to induce expression of pro-inflammatory molecules was diminished in comparison to that of the LPS [18]. The present study expands our previous observations to LOS from an NTHi strain that was isolated from a pediatric case of otitis media [20] indicating that the capacity of H. influenzae LOS to activate innate immune cells is not restricted to particular serotypes of the pathogen.
It is well established that NTHi bacteria are distinct and more genetically variable compared to encapsulated H. influenzae, and considerable heterogeneity exists among the structures of lipid A expressed by different H. influenzae strains [36, 37]. Because such heterogeneity may be manifested in differentially acylated lipid A forms that would determine the interaction with the TLR4 receptor complex, it may potentially result in different degrees of immunostimulatory ability among the different strains. It is interesting that despite the great genetic diversity among H. influenzae strains, LOS of NTHi shares immunostimulatory capacity with LOS isolated from Eagan and Rd [18].
In fact, LOS of NTHi strain 375, similarly to LOS of Eagan and Rd, had a marked ability to induce the expression of both MHC class I and II antigen-presenting molecules that was similar to those of E. coli LPS. Moreover, we found that NTHi LOS induced higher expression of HLA-DR as compared to the corresponding numbers of killed NTHi. This may be explained by different effects of whole bacteria versus isolated LOS on the production of the anti-inflammatory cytokine IL-10. Differentiated THP-1 cells that acquired macrophage-like properties produced lower amounts of IL-10 when stimulated with LOS as compared to the corresponding numbers of whole bacteria. This is not surprising because production of IL-10 is regulated by signaling initiated by the engagement of several pattern-recognition receptors including TLR2, TLR4, TLR9, and Nod-like receptors [38]. Because IL-10 is known to down-regulate the cellular expression of MHC molecules [39], its higher production induced by whole NTHi may explain differences in MHC class II expression induced by LOS versus NTHi. Of note, IFN-γ which is the major cytokine upregulating the MHC class II expression on monocytes [40] was not induced by either whole killed NTHi or LOS. Indeed, production of IFN-γ by human macrophages could require additional signals, such as IL-12 in combination with IL-18 [41]. Although using killed bacteria in an experimental model does not reproduce the full spectrum of biological events triggered by NTHi in vivo, our approach can nevertheless be clinically relevant. In cases of NTHi infections treated with antibiotics, the immune system is exposed to large numbers of killed bacteria, and hence cellular responses studied in our model may reflect actual activation of innate immune cells, including both immature monocytes and macrophage-like differentiated monocytic cells.
The results of this study suggest that isolated LOS of NTHi may represent
an attractive vaccine candidate to prevent invasive disease caused by this
pathogen. Importantly, isolated LOS had decreased pro-inflammatory ability
as compared to typical LPS of gram-negative bacteria or whole NTHi
bacteria, specifically in terms of TNF-
This work was supported by a National Science and Engineering Research Council (NSERC) Discovery Grant and Northern Ontario School of Medicine (NOSM) Faculty Association Research Development Award to M.U. We would also like to thank Dr. Andrew Cox and Frank St. Michael for their kind donation of the 375 LOS.
The authors do not have a conflict to declare.
1. Van Eldere J, Slack MP, Ladhani S, Cripps AW. Non-typeable Haemophilus influenzae, an under-recognised pathogen. Lancet Infect Dis. 2014;14(12):1281-92. PubMed PMID: 25012226. doi: 10.1016/S1473-3099(14)70734-0
2. Klein JO. Role of nontypeable Haemophilus influenzae in pediatric respiratory tract infections. Pediatr Infect Dis J. 1997;16(2 Suppl):S5-8. PubMed PMID: 9041620.
3. van Wessel K, Rodenburg GD, Veenhoven RH, Spanjaard L, van der Ende A, Sanders EA. Nontypeable Haemophilus influenzae invasive disease in The Netherlands: a retrospective surveillance study 2001-2008. Clin Infect Dis. 2011;53(1):e1-7. PubMed PMID: 21653293. doi: 10.1093/cid/cir268
4. Ladhani S, Slack MP, Heath PT, von Gottberg A, Chandra M, Ramsay ME, European Union Invasive Bacterial Infection Surveillance participants. Invasive Haemophilus influenzae Disease, Europe, 1996-2006. Emerg Infect Dis. 2010;16(3):455-63. PubMed PMID: 20202421. Pubmed Central PMCID: PMC3322004. doi: 10.3201/eid1603.090290
5. Dworkin MS, Park L, Borchardt SM. The changing epidemiology of invasive Haemophilus influenzae disease, especially in persons > or = 65 years old. Clin Infect Dis. 2007;44(6):810-6. PubMed PMID: 17304452. doi: 10.1086/511861
6. Prymula R, Peeters P, Chrobok V, Kriz P, Novakova E, Kaliskova E, Kohl I, Lommel P, Poolman J, Prieels JP, Schuerman L. Pneumococcal capsular polysaccharides conjugated to protein D for prevention of acute otitis media caused by both Streptococcus pneumoniae and non-typable Haemophilus influenzae: a randomised double-blind efficacy study. Lancet. 2006;367(9512):740-8. PubMed PMID: 16517274. doi: 10.1016/S0140-6736(06)68304-9
7. Murphy TF, Nelson MB, Dudas KC, Mylotte JM, Apicella MA. Identification of a specific epitope of Haemophilus influenzae on a 16,600-dalton outer membrane protein. J Infect Dis. 1985;152(6):1300-7. PubMed PMID: 2415644.
8. Nelson MB, Munson RS, Jr., Apicella MA, Sikkema DJ, Molleston JP, Murphy TF. Molecular conservation of the P6 outer membrane protein among strains of Haemophilus influenzae: analysis of antigenic determinants, gene sequences, and restriction fragment length polymorphisms. Infect Immun. 1991;59(8):2658-63. PubMed PMID: 1713197. Pubmed Central PMCID: PMC258070.
9. Forsgren A, Riesbeck K, Janson H. Protein D of Haemophilus influenzae: a protective nontypeable H. influenzae antigen and a carrier for pneumococcal conjugate vaccines. Clin Infect Dis. 2008;46(5):726-31. PubMed PMID: 18230042. doi: 10.1086/527396
10. Prasadarao NV, Lysenko E, Wass CA, Kim KS, Weiser JN. Opacity-associated protein A contributes to the binding of Haemophilus influenzae to chang epithelial cells. Infect Immun. 1999;67(8):4153-60. PubMed PMID: 10417187. Pubmed Central PMCID: PMC96720.
11. Poulsen K, Brandt J, Hjorth JP, Thogersen HC, Kilian M. Cloning and sequencing of the immunoglobulin A1 protease gene (iga) of Haemophilus influenzae serotype b. Infect Immun. 1989;57(10):3097-105. PubMed PMID: 2506130. Pubmed Central PMCID: PMC260775.
12. Gu XX, Rudy SF, Chu C, McCullagh L, Kim HN, Chen J, Li J, Robbins JB, Van Waes C, Battey JF. Phase I study of a lipooligosaccharide-based conjugate vaccine against nontypeable Haemophilus influenzae. Vaccine. 2003;21(17-18):2107-14. PubMed PMID: 12706701.
13. Schweda EK, Richards JC, Hood DW, Moxon ER. Expression and structural diversity of the lipopolysaccharide of Haemophilus influenzae: implication in virulence. Int J Med Microbiol. 2007;297(5):297-306. PubMed PMID: 17452015. doi: 10.1016/j.ijmm.2007.03.007
14. Triantafilou M, Triantafilou K. Lipopolysaccharide recognition: CD14, TLRs and the LPS-activation cluster. Trends Immunol. 2002;23(6):301-4. PubMed PMID: 12072369.
15. Paciello I, Silipo A, Lembo-Fazio L, Curcuru L, Zumsteg A, Noel G, Ciancarella V, Sturiale L, Molinaro A, Bernardini ML. Intracellular Shigella remodels its LPS to dampen the innate immune recognition and evade inflammasome activation. Proc Natl Acad Sci U S A. 2013;110(46):E4345-54. PubMed PMID: 24167293. Pubmed Central PMCID: PMC3832022. doi: 10.1073/pnas.1303641110
16. Morey P, Viadas C, Euba B, Hood DW, Barberan M, Gil C, Grillo MJ, Bengoechea JA, Garmendia J. Relative contributions of lipooligosaccharide inner and outer core modifications to nontypeable Haemophilus influenzae pathogenesis. Infect Immun. 2013;81(11):4100-11. PubMed PMID: 23980106. Pubmed Central PMCID: PMC3811809. doi: 10.1128/IAI.00492-13
17. Mansson M, Bauer SH, Hood DW, Richards JC, Moxon ER, Schweda EK. A new structural type for Haemophilus influenzae lipopolysaccharide. Structural analysis of the lipopolysaccharide from nontypeable Haemophilus influenzae strain 486. Eur J Biochem. 2001;268(7):2148-59. PubMed PMID: 11277939.
18. Choi J, Cox AD, Li J, McCready W, Ulanova M. Activation of innate immune responses by Haemophilus influenzae lipooligosaccharide. Clin Vaccine Immunol. 2014;21(5):769-76. PubMed PMID: 24671554. Pubmed Central PMCID: PMC4018885. doi: 10.1128/CVI.00063-14
19. Choi J, Nix EB, Gaultier GN, Cox AD, McCready W, Ulanova M. Naturally occurring bactericidal antibodies specific for Haemophilus influenzae lipooligosaccharide are present in healthy adult individuals. Vaccine. 2015;33(16):1941-7. PubMed PMID: 25738817. doi: 10.1016/j.vaccine.2015.02.060
20. Mell JC, Sinha S, Balashov S, Viadas C, Grassa CJ, Ehrlich GD, Nislow C, Redfield RJ, Garmendia J. Complete Genome Sequence of Haemophilus influenzae Strain 375 from the Middle Ear of a Pediatric Patient with Otitis Media. Genome Announc. 2014;2(6). PubMed PMID: 25477405. Pubmed Central PMCID: PMC4256186. doi: 10.1128/genomeA.01245-14
21. Gu XX, Tsai CM, Apicella MA, Lim DJ. Quantitation and biological properties of released and cell-bound lipooligosaccharides from nontypeable Haemophilus influenzae. Infect Immun. 1995;63(10):4115-20. PubMed PMID: 7558327. Pubmed Central PMCID: PMC173578.
22. Wang X, Moser C, Louboutin JP, Lysenko ES, Weiner DJ, Weiser JN, Wilson JM. Toll-like receptor 4 mediates innate immune responses to Haemophilus influenzae infection in mouse lung. J Immunol. 2002;168(2):810-5. PubMed PMID: 11777976.
23. Lee MS, Kim YJ. Signaling pathways downstream of pattern-recognition receptors and their cross talk. Annu Rev Biochem. 2007;76:447-80. PubMed PMID: 17328678. 10.1146/annurev.biochem.doi: 76.060605.122847
24. Liu SF, Malik AB. NF-kappa B activation as a pathological mechanism of septic shock and inflammation. Am J Physiol Lung Cell Mol Physiol. 2006;290(4):L622-L45. PubMed PMID: 16531564. doi: 10.1152/ajplung.00477.2005
25. McDonald C, Inohara N, Nunez G. Peptidoglycan signaling in innate immunity and inflammatory disease. J Biol Chem. 2005;280(21):20177-80. PubMed PMID: 15802263. doi: 10.1074/jbc.R500001200
26. Pauleau AL, Murray PJ. Role of nod2 in the response of macrophages to toll-like receptor agonists. Mol Cell Biol. 2003;23(21):7531-9. PubMed PMID: 14560001. Pubmed Central PMCID: PMC207570.
27. Tsai WH, Huang DY, Yu YH, Chen CY, Lin WW. Dual roles of NOD2 in TLR4-mediated signal transduction and -induced inflammatory gene expression in macrophages. Cell Microbiol. 2011;13(5):717-30. PubMed PMID: 21199260. doi: 10.1111/j.1462-5822.2010.01567.x
28. Takahashi Y, Isuzugawa K, Murase Y, Imai M, Yamamoto S, Iizuka M, Akira S, Bahr GM, Momotani E, Hori M, Ozaki H, Imakawa K. Up-regulation of NOD1 and NOD2 through TLR4 and TNF-alpha in LPS-treated murine macrophages. J Vet Med Sci. 2006;68(5):471-8. PubMed PMID: 16757890.
29. Shuto T, Xu H, Wang B, Han J, Kai H, Gu XX, Murphy TF, Lim DJ, Li JD. Activation of NF-kappa B by nontypeable Hemophilus influenzae is mediated by toll-like receptor 2-TAK1-dependent NIK-IKK alpha /beta-I kappa B alpha and MKK3/6-p38 MAP kinase signaling pathways in epithelial cells. Proc Natl Acad Sci U S A. 2001;98(15):8774-9. PubMed PMID: 11438700. Pubmed Central PMCID: PMC37511. doi: 10.1073/pnas.151236098
30. Galdiero M, Galdiero M, Finamore E, Rossano F, Gambuzza M, Catania MR, Teti G, Midiri A, Mancuso G. Haemophilus influenzae porin induces Toll-like receptor 2-mediated cytokine production in human monocytes and mouse macrophages. Infect Immun. 2004;72(2):1204-9. PubMed PMID: 14742577. Pubmed Central PMCID: PMC321594.
31. Punturieri A, Copper P, Polak T, Christensen PJ, Curtis JL. Conserved nontypeable Haemophilus influenzae-derived TLR2-binding lipopeptides synergize with IFN-beta to increase cytokine production by resident murine and human alveolar macrophages. J Immunol. 2006;177(1):673-80. PubMed PMID: 16785566. Pubmed Central PMCID: PMC2373263.
32. Lugade AA, Bianchi-Smiraglia A, Pradhan V, Elkin G, Murphy TF, Thanavala Y. Lipid motif of a bacterial antigen mediates immune responses via TLR2 signaling. PLoS One. 2011;6(5):e19781. PubMed PMID: 21611194. Pubmed Central PMCID: PMC3096640. doi: 10.1371/journal.pone.0019781
33. Leichtle A, Hernandez M, Lee J, Pak K, Webster NJ, Wollenberg B, Wasserman SI, Ryan AF. The role of DNA sensing and innate immune receptor TLR9 in otitis media. Innate Immun. 2012;18(1):3-13. PubMed PMID: 21239460. Pubmed Central PMCID: PMC4041324. doi: 10.1177/1753425910393539
34. Rotta Detto Loria J, Rohmann K, Droemann D, Kujath P, Rupp J, Goldmann T, Dalhoff K. Nontypeable Haemophilus Influenzae Infection Upregulates the NLRP3 Inflammasome and Leads to Caspase-1-Dependent Secretion of Interleukin-1beta - A Possible Pathway of Exacerbations in COPD. PLoS One. 2013;8(6):e66818. PubMed PMID: 23840534. Pubmed Central PMCID: PMC3694113. doi: 10.1371/journal.pone.0066818
35. Ng GZ, Sutton P. The MUC1 mucin specifically inhibits activation of the NLRP3 inflammasome. Genes Immun. 2016;17(3):203-6. PubMed PMID: 26938663. doi: 10.1038/gene.2016.10
36. Swords WE, Jones PA, Apicella MA. The lipo-oligosaccharides of Haemophilus influenzae: an interesting array of characters. J Endotoxin Res. 2003;9(3):131-44. PubMed PMID: 12831454. doi: 10.1179/096805103125001531
37. Apicella MA, Dudas KC, Campagnari A, Rice P, Mylotte JM, Murphy TF. Antigenic heterogeneity of lipid A of Haemophilus influenzae. Infect Immun. 1985;50(1):9-14. PubMed PMID: 3899941. Pubmed Central PMCID: PMC262125.
38. Saraiva M, O'Garra A. The regulation of IL-10 production by immune cells. Nat Rev Immunol. 2010;10(3):170-81. PubMed PMID: 20154735. doi: 10.1038/nri2711
39. Thomssen H, Kahan M, Londei M. Differential effects of interleukin-10 on the expression of HLA class II and CD1 molecules induced by granulocyte/macrophage colony-stimulating factor/interleukin-4. Eur J Immunol. 1995;25(9):2465-70. PubMed PMID: 7589112. doi: 10.1002/eji.1830250909
40. Collins T, Korman AJ, Wake CT, Boss JM, Kappes DJ, Fiers W, Ault KA, Gimbrone MA, Jr., Strominger JL, Pober JS. Immune interferon activates multiple class II major histocompatibility complex genes and the associated invariant chain gene in human endothelial cells and dermal fibroblasts. Proc Natl Acad Sci U S A. 1984;81(15):4917-21. PubMed PMID: 6431411. Pubmed Central PMCID: PMC391603.
41. Darwich L, Coma G, Pena R, Bellido R, Blanco EJ, Este JA, Borras FE, Clotet B, Ruiz L, Rosell A, Andreo F, Parkhouse RM, Bofill M. Secretion of interferon-gamma by human macrophages demonstrated at the single-cell level after costimulation with interleukin (IL)-12 plus IL-18. Immunology. 2009;126(3):386-93. PubMed PMID: 18759749. Pubmed Central PMCID: PMC2669819. doi: 10.1111/j.1365-2567.2008.02905.x